
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

You can install CodelistGenerator from CRAN
install.packages("CodelistGenerator")Or you can also install the development version of CodelistGenerator
install.packages("remotes")
remotes::install_github("darwin-eu/CodelistGenerator")library(dplyr)
library(CDMConnector)
library(CodelistGenerator)For this example we’ll use the Eunomia dataset (which only contains a subset of the OMOP CDM vocabularies)
requireEunomia()
db <- DBI::dbConnect(duckdb::duckdb(), dbdir = eunomiaDir())
cdm <- cdmFromCon(db,
cdmSchema = "main",
writeSchema = "main",
writePrefix = "cg_")OMOP CDM vocabularies are frequently updated, and we can identify the version of the vocabulary of our Eunomia data
vocabularyVersion(cdm = cdm)
#> [1] "v5.0 18-JAN-19"CodelistGenerator provides functions to extract code lists based on vocabulary hierarchies. One example is `getDrugIngredientCodes, which we can use, for example, to get the concept IDs used to represent aspirin and diclofenac.
ing <- getDrugIngredientCodes(cdm = cdm,
name = c("aspirin", "diclofenac"),
nameStyle = "{concept_name}")
ing
#>
#> - aspirin (2 codes)
#> - diclofenac (1 codes)
ing$aspirin
#> [1] 1112807 19059056
ing$diclofenac
#> [1] 1124300CodelistGenerator can also support systematic searches of the vocabulary tables to support codelist development. A little like the process for a systematic review, the idea is that for a specified search strategy, CodelistGenerator will identify a set of concepts that may be relevant, with these then being screened to remove any irrelevant codes by clinical experts.
We can do a simple search for asthma
asthma_codes1 <- getCandidateCodes(
cdm = cdm,
keywords = "asthma",
domains = "Condition"
)
asthma_codes1 |>
glimpse()
#> Rows: 2
#> Columns: 6
#> $ concept_id <int> 317009, 4051466
#> $ found_from <chr> "From initial search", "From initial search"
#> $ concept_name <chr> "Asthma", "Childhood asthma"
#> $ domain_id <chr> "Condition", "Condition"
#> $ vocabulary_id <chr> "SNOMED", "SNOMED"
#> $ standard_concept <chr> "S", "S"But perhaps we want to exclude certain concepts as part of the search strategy, in this case we can add these like so
asthma_codes2 <- getCandidateCodes(
cdm = cdm,
keywords = "asthma",
exclude = "childhood",
domains = "Condition"
)
asthma_codes2 |>
glimpse()
#> Rows: 1
#> Columns: 6
#> $ concept_id <int> 317009
#> $ found_from <chr> "From initial search"
#> $ concept_name <chr> "Asthma"
#> $ domain_id <chr> "Condition"
#> $ vocabulary_id <chr> "SNOMED"
#> $ standard_concept <chr> "S"As well as functions for finding codes, we also have functions to summarise their use. Here for
library(flextable)
asthma_code_use <- summariseCodeUse(list("asthma" = asthma_codes1$concept_id) |>
newCodelist(),
cdm = cdm
)
tableCodeUse(asthma_code_use, type = "flextable", style = "darwin")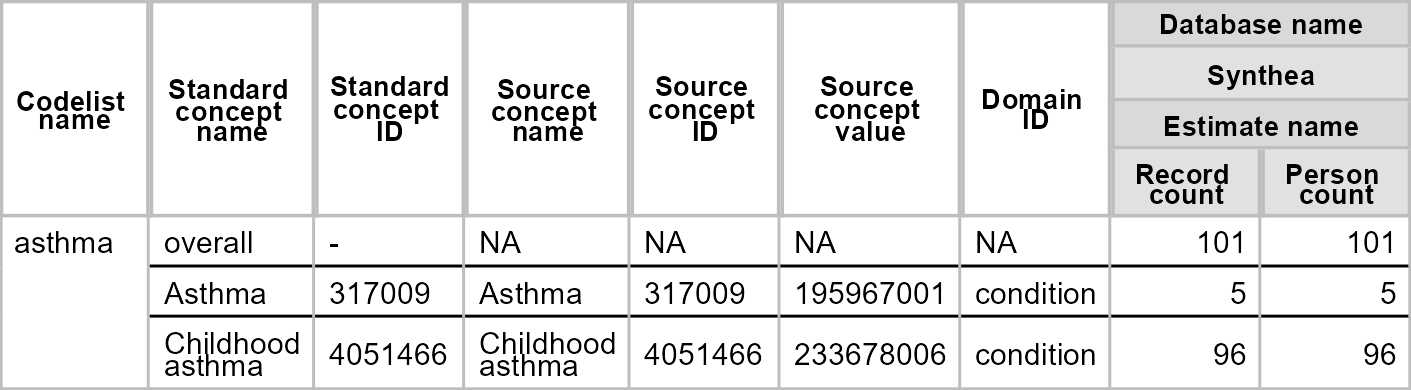
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.