
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

Supporting functionality to run ‘caret’ with spatial or spatial-temporal data. ‘caret’ is a frequently used package for model training and prediction using machine learning. CAST includes functions to improve spatial or spatial-temporal modelling tasks using ‘caret’. To decrease spatial overfitting and to improve model performances, the package implements a forward feature selection that selects suitable predictor variables in view to their contribution to spatial or spatio-temporal model performance. CAST further includes functionality to estimate the (spatial) area of applicability of prediction models.
Note: The developer version of CAST can be found on https://github.com/HannaMeyer/CAST. The CRAN Version can be found on https://CRAN.R-project.org/package=CAST
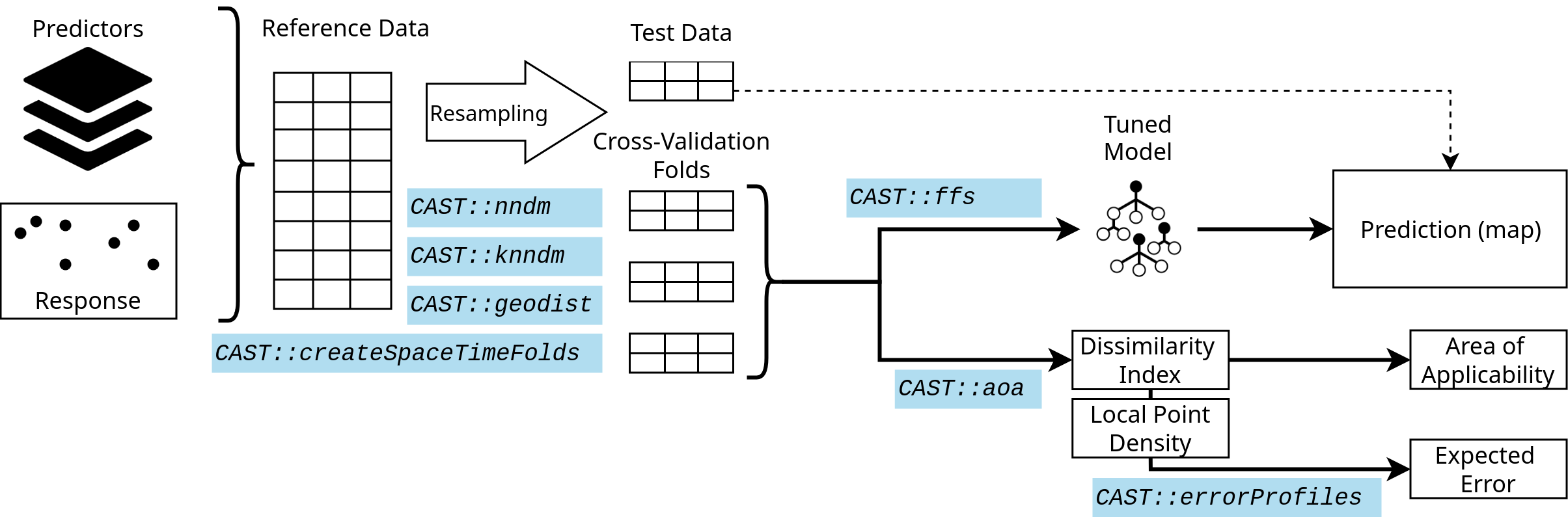 The figure shows a very simple workflow for a spatial prediction
mapping workflow, indicating which function in CAST can be used in the
different steps to support the spatial prediction.
The figure shows a very simple workflow for a spatial prediction
mapping workflow, indicating which function in CAST can be used in the
different steps to support the spatial prediction.
https://hannameyer.github.io/CAST/
Book chapter: The CAST package for training and assessment of spatial prediction models in R. Preprint
The talk from the OpenGeoHub summer school 2019 on spatial validation and variable selection: https://www.youtube.com/watch?v=mkHlmYEzsVQ.
Tutorial (https://youtu.be/EyP04zLe9qo) and Lecture (https://youtu.be/OoNH6Nl-X2s) recording from OpenGeoHub summer school 2020 on the area of applicability. As well as talk at the OpenGeoHub summer school 2021: https://av.tib.eu/media/54879
Talk and tutorial from the OpenGeoHub 2022 summer school on Machine learning-based maps of the environment - challenges of extrapolation and overfitting, including discussions on the area of applicability and the nearest neighbor distance matching cross-validation (https://doi.org/10.5446/59412).
Milà, C., Mateu, J., Pebesma, E., Meyer, H. (2022): Nearest Neighbour Distance Matching Leave-One-Out Cross-Validation for map validation. Methods in Ecology and Evolution 13, 1304– 1316. https://doi.org/10.1111/2041-210X.13851
Linnenbrink, J., Milà, C., Ludwig, M., and Meyer, H.: kNNDM (2024): k-fold Nearest Neighbour Distance Matching Cross-Validation for map accuracy estimation. Geosci. Model Dev., 17, 5897–5912. https://doi.org/10.5194/gmd-17-5897-2024.
Meyer, H., Reudenbach, C., Hengl, T., Katurji, M., Nauss, T. (2018): Improving performance of spatio-temporal machine learning models using forward feature selection and target-oriented validation. Environmental Modelling & Software, 101, 1-9. https://doi.org/10.1016/j.envsoft.2017.12.001
Meyer, H., Reudenbach, C., Hengl, T., Katurji, M., Nauss, T. (2018): Improving performance of spatio-temporal machine learning models using forward feature selection and target-oriented validation. Environmental Modelling & Software, 101, 1-9. https://doi.org/10.1016/j.envsoft.2017.12.001
Meyer, H., Reudenbach, C., Wöllauer, S., Nauss, T. (2019): Importance of spatial predictor variable selection in machine learning applications - Moving from data reproduction to spatial prediction. Ecological Modelling. 411. https://doi.org/10.1016/j.ecolmodel.2019.108815
Meyer, H., Pebesma, E. (2021). Predicting into unknown space? Estimating the area of applicability of spatial prediction models. Methods in Ecology and Evolution, 12, 1620– 1633. https://doi.org/10.1111/2041-210X.13650
Schumacher, F., Knoth, C., Ludwig, M., Meyer, H. (2025): Estimation of local training data point densities to support the assessment of spatial prediction uncertainty. Geosci. Model Dev., 18, 10185–10202. https://doi.org/10.5194/gmd-18-10185-2025.
Meyer, H., Pebesma, E. (2022): Machine learning-based global maps of ecological variables and the challenge of assessing them. Nature Communications, 13. https://www.nature.com/articles/s41467-022-29838-9
Ludwig, M., Moreno-Martinez, A., Hoelzel, N., Pebesma, E., Meyer, H. (2023): Assessing and improving the transferability of current global spatial prediction models. Global Ecology and Biogeography, 32, 356–368. https://doi.org/10.1111/geb.13635.
Milà, C., Ludwig, M., Pebesma, E., Tonne, C., and Meyer, H. (2024): Random forests with spatial proxies for environmental modelling: opportunities and pitfalls. Geosci. Model Dev., 17, 6007–6033. https://doi.org/10.5194/gmd-17-6007-2024.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.