The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The goal of BioVizSeq is to visualize the types and distribution of elements within bio-sequences. At the same time, We have developed a geom layer, geom_rrect(), that can generate rounded rectangles. No external references are used in the development of this package.
Install from CRAN:
# Install from CRAN
install.packages("BioVizSeq")Install from Github: the development version of BioVizSeq:
install.packages("devtools")
devtools::install_github("zhaosq2022/BioVizSeq")library(BioVizSeq)
#> Registered S3 methods overwritten by 'treeio':
#> method from
#> MRCA.phylo tidytree
#> MRCA.treedata tidytree
#> Nnode.treedata tidytree
#> Ntip.treedata tidytree
#> ancestor.phylo tidytree
#> ancestor.treedata tidytree
#> child.phylo tidytree
#> child.treedata tidytree
#> full_join.phylo tidytree
#> full_join.treedata tidytree
#> groupClade.phylo tidytree
#> groupClade.treedata tidytree
#> groupOTU.phylo tidytree
#> groupOTU.treedata tidytree
#> inner_join.phylo tidytree
#> inner_join.treedata tidytree
#> is.rooted.treedata tidytree
#> nodeid.phylo tidytree
#> nodeid.treedata tidytree
#> nodelab.phylo tidytree
#> nodelab.treedata tidytree
#> offspring.phylo tidytree
#> offspring.treedata tidytree
#> parent.phylo tidytree
#> parent.treedata tidytree
#> root.treedata tidytree
#> rootnode.phylo tidytree
#> sibling.phylo tidytree
#> Package BioVizSeq loaded successfully!
# Extra package
library(ggplot2)
#> Warning: 程辑包'ggplot2'是用R版本4.3.3 来建造的gff or gtf file
gff_path <- system.file("extdata", "idpro.gff3", package = "BioVizSeq")
gff_data <- read.table(gff_path, header = FALSE, sep = '\t')
gff_loc <- gff_to_loc(gff_data)
motif_plot(gff_loc$table_loc, gff_loc$gene_length) +
labs(x="DNA length (5'-3')", y="Gene name")
gff_path <- system.file("extdata", "idpro.gff3", package = "BioVizSeq")
gff_plot(gff_path)
meme.xml or mast.xml
meme_path <- system.file("extdata", "mast.xml", package = "BioVizSeq")
meme_file <- readLines(meme_path)
motif_loc <- meme_to_loc(meme_file)
motif_plot(motif_loc$table_loc, motif_loc$gene_length)
meme_path <- system.file("extdata", "mast.xml", package = "BioVizSeq")
meme_plot(meme_path)
Download: .tsv
pfam_path <- system.file("extdata", "iprscan.tsv", package = "BioVizSeq")
pfam_file <- read.table(pfam_path, sep='\t', header = FALSE)
domain_loc <- pfam_to_loc(pfam_file)
motif_plot(domain_loc$table_loc, domain_loc$gene_length)
pfam_path <- system.file("extdata", "iprscan.tsv", package = "BioVizSeq")
pfam_plot(pfam_path)
Download “Superfamily Only”
Type: .txt
hitdata_path <- system.file("extdata", "hitdata.txt", package = "BioVizSeq")
cdd_file <- readLines(hitdata_path)
domain_loc <- cdd_to_loc(cdd_file)
fa_path <- system.file("extdata", "idpep.fa", package = "BioVizSeq")
gene_length <- fastaleng(fa_path)
motif_plot(domain_loc, gene_length)
hitdata_path <- system.file("extdata", "hitdata.txt", package = "BioVizSeq")
fa_path <- system.file("extdata", "idpep.fa", package = "BioVizSeq")
cdd_plot(hitdata_path, fa_path)
protein file (.fa or .fasta)
fa_path <- system.file("extdata", "target.fa", package = "BioVizSeq")
domain_loc <- smart_to_loc(fa_path)
#> Submitting sequence AtAP2_002...
#> Submitting sequence AtAP2_003...
#> Job entered the queue with ID12315310532459281744966748fjuQJesKfo. Waiting for results.
#> Submitting sequence AtAP2_004...
#> Submitting sequence AtAP2_005...
motif_plot(domain_loc$table_loc, domain_loc$gene_length)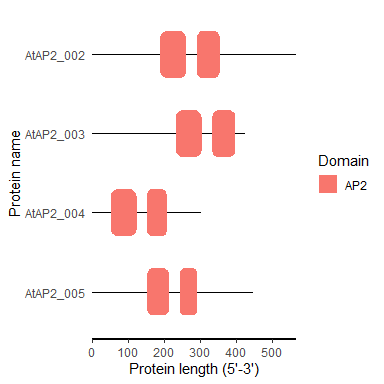
fa_path <- system.file("extdata", "target.fa", package = "BioVizSeq")
smart_plot(fa_path)
#> Submitting sequence AtAP2_002...
#> Submitting sequence AtAP2_003...
#> Job entered the queue with ID12315310532468761744966784YObRQLBBcV. Waiting for results.
#> Submitting sequence AtAP2_004...
#> Submitting sequence AtAP2_005...
promoter sequence(.fa or .fasta)
# 1. upload fasta file to plantcare, get the result file(.tab)
# upload_fa_to_plantcare(fasta_file, email)
# 2. Classify the functions of cis element
plantcare_path <- system.file("extdata", "plantCARE_output.tab", package = "BioVizSeq")
plantcare_file <- read.table(plantcare_path, header = FALSE, sep = '\t', quote="")
plantcare_data <- plantcare_classify(plantcare_file)
plantcare_loc <- plantcare_to_loc(plantcare_data)
promoter_length <- data.frame(ID = unique(plantcare_loc$ID), length=2000)
motif_plot(plantcare_loc, promoter_length) +
labs(x="Promoter Length", y="Gene")
plantcare_path <- system.file("extdata", "plantCARE_output.tab", package = "BioVizSeq")
plantcare_plot(plantcare_path, promoter_length = 2000)
p_tree, p_gff, p_pfam, p_meme, p_smart, p_cdd, p_plantcare
library(patchwork)
tree_path <- system.file("extdata", "idpep.nwk", package = "BioVizSeq")
gff_path <- system.file("extdata", "idpro.gff3", package = "BioVizSeq")
meme_path <- system.file("extdata", "mast.xml", package = "BioVizSeq")
pfam_path <- system.file("extdata", "iprscan.tsv", package = "BioVizSeq")
plot_file <- combi_p(tree_path = tree_path, gff_path = gff_path,
meme_path = meme_path, pfam_path = pfam_path)
plot_file$p_tree + plot_file$p_gff + plot_file$p_pfam +
plot_file$p_meme +plot_layout(ncol = 4, guides = 'collect') +
plot_annotation(
tag_levels = 'A'
)
library(patchwork)
tree_path <- system.file("extdata", "idpep.nwk", package = "BioVizSeq")
plantcare_path <- system.file("extdata", "plantCARE_output.tab", package = "BioVizSeq")
plot_file <- combi_p(tree_path = tree_path, plantcare_path = plantcare_path, promoter_length = 2000)
plot_file$p_tree + plot_file$p_plantcare1 + plot_file$p_plantcare2 + plot_layout(ncol = 3, guides = 'collect', widths = c(1, 3, 1)) + plot_annotation( tag_levels = 'A' )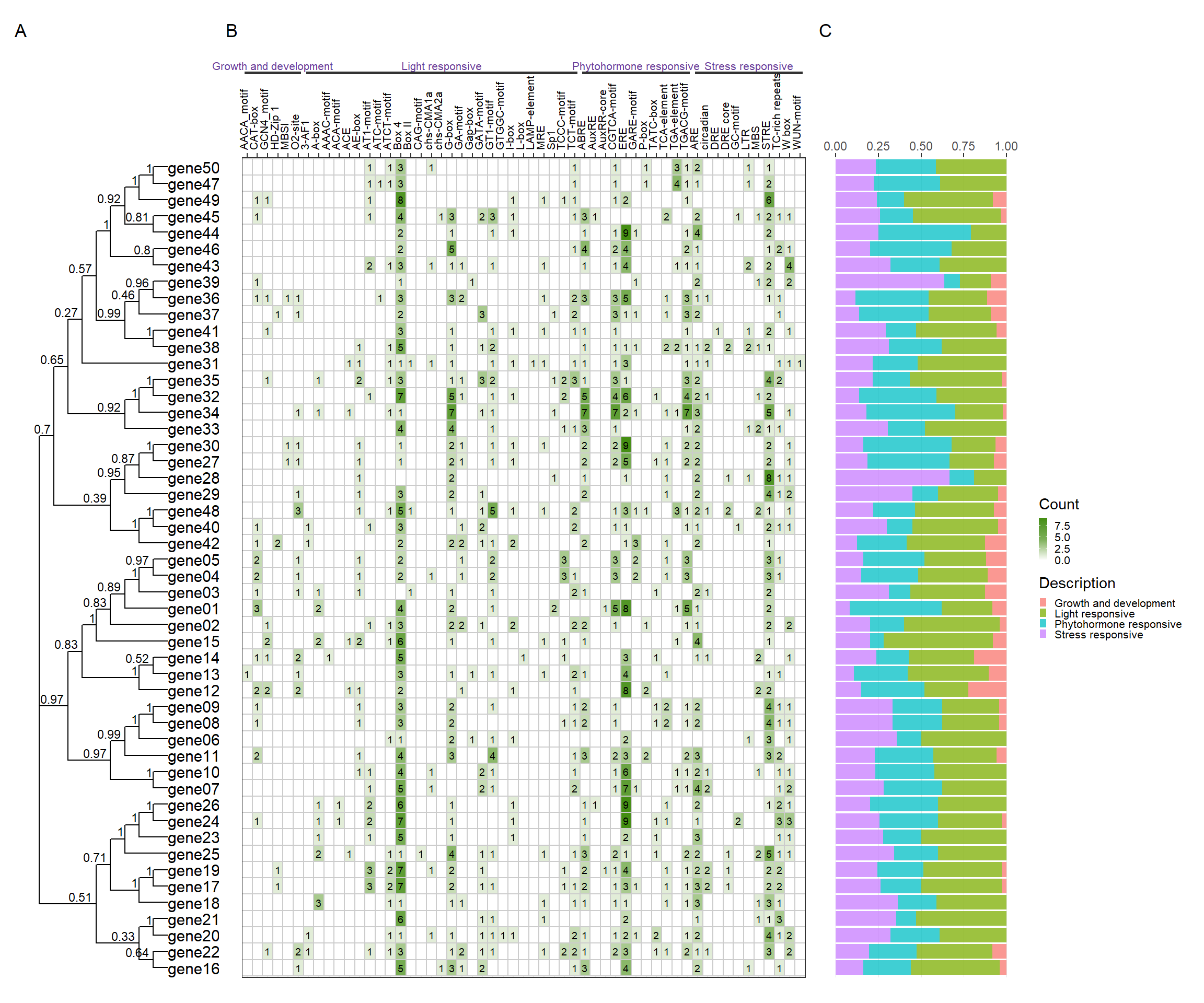
gff_path <- system.file("extdata", "idpro.gff3", package = "BioVizSeq")
gff_data <- read.table(gff_path, header = FALSE, sep = '\t')
gene_statistics_data <- gff_statistics(gff_data)
head(gene_statistics_data)
#> ID Location Chain gene_length CDS_length protein_length
#> 1 gene01 Chr15:31085288-31086321 - 1034 531 176
#> 2 gene02 Contig862:15967-16631 - 665 555 184
#> 3 gene03 Chr15:31004816-31005518 + 703 564 187
#> 4 gene04 Chr15:30780257-30780955 + 699 564 187
#> 5 gene05 Chr15:30976079-30976776 + 698 564 187
#> 6 gene06 Chr2:12719447-12720989 + 1543 1224 407
#> exon_number intron_number CDS_number UTR_number
#> 1 2 1 2 2
#> 2 2 1 2 0
#> 3 2 1 2 0
#> 4 2 1 2 0
#> 5 2 1 2 0
#> 6 1 0 1 2
pep_path <- system.file("extdata", "idpep2.fa", package = "BioVizSeq")
pep_calc_result <- ProtParam_calc(pep_path)
#> Submitting sequence gene01...
#> Submitting sequence gene02...
#> Submitting sequence gene03...
head(pep_calc_result)
#> ID Number of amino acids Molecular weight Theoretical pI
#> 1 gene01 176 19433.92 6.22
#> 2 gene02 184 20288.83 9.07
#> 3 gene03 187 21042.90 7.68
#> The instability index Aliphatic index Grand average of hydropathicity
#> 1 80.30 67.16 -0.611
#> 2 68.69 73.15 -0.580
#> 3 72.86 69.41 -0.637These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.