The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
BAQM supplies functions developed by Babson College instructors for AQM 1000 and AQM 2000 courses using R in the curriculum. The primary functions provide:
sumry.df() - summary descriptive statistics for data
frames, allowing both numeric and factor variables, in both wide and
long formats.sumry.lm() - expanded statistics and enhanced
formatting to summarize linear model results.lm_plot.4way() - multiple diagnostic plots for linear
models using ggplot, including a 4-in-1 summary graphic.sumry.regsubsets() - compact “best subsets” linear
model reports for analytics from the regsubsets function of
the leaps package.You can install the development version of BAQM from GitHub with:
install.packages("pak")
pak::pak("CPA-wrk/BAQM")These examples use the built-in R data sets iris,
swiss, and mtcars, and show:
iris and
swiss,swiss,iris and miles per gallon in mtcars, andmtcars model.(Variable names are truncated in swiss to narrow the
output.)
library(leaps)
library(BAQM)
#
sumry(iris) # Includes non-numeric variable
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> n.val 150 150 150 150 150
#> n.na 0 0 0 0 0
#> min 4.3 2 1 0.1 n.lvl : 3
#> Q1 5.1 2.7 1.6 0.2 setosa :50
#> median 5.8 3 4.35 1.3 versclr:50
#> mean 5.843 3.057 3.758 1.199 virginc:50
#> Q3 6.45 3.4 5.1 1.8
#> max 7.9 4.4 6.9 2.5
#> std.dev 0.8281 0.4359 1.765 0.7622
#
names(swiss) # Show original variable names
#> [1] "Fertility" "Agriculture" "Examination" "Education"
#> [5] "Catholic" "Infant.Mortality"
names(swiss) <- substr(names(swiss), 1, 4) # Narrows output
sumry(swiss)
#> Fert Agri Exam Educ Cath Infa
#> n.val 47 47 47 47 47 47
#> n.na 0 0 0 0 0 0
#> min 35 1.2 3 1 2.15 10.8
#> Q1 64.4 35.3 12 6 5.16 18
#> median 70.4 54.1 16 8 15.14 20
#> mean 70.14 50.66 16.49 10.98 41.14 19.94
#> Q3 79.3 67.8 22 12.5 93.4 22.2
#> max 92.5 89.7 37 53 100 26.6
#> std.dev 12.49 22.71 7.978 9.615 41.7 2.913
regs <- regsubsets(Fert ~ ., data = swiss, nbest = 3)
sumry(regs)
#>
#> Call: (function (...)
#> rmarkdown::render(...))(input = base::quote("/Users/peter/Library/CloudStorage/OneDrive-centerpointanalytics.com/CPA_wrk/R/BAQM/README.Rmd"),
#> output_options = base::quote(list(html_preview = FALSE)),
#> quiet = base::quote(TRUE))
#> _k_i.best rsq adjr2 see cp Agri Exam Educ Cath Infa
#> 1 1 ( 1 ) 0.4406 0.4282 9.446029 35.20 *
#> 2 1 ( 2 ) 0.4172 0.4042 9.642000 38.48 *
#> 3 1 ( 3 ) 0.2150 0.1976 11.189945 66.75 *
#> 4 2 ( 1 ) 0.5745 0.5552 8.331442 18.49 * *
#> 5 2 ( 2 ) 0.5648 0.5450 8.426136 19.85 * *
#> 6 2 ( 3 ) 0.5363 0.5152 8.697447 23.83 * *
#> 7 3 ( 1 ) 0.6625 0.6390 7.505417 8.18 * * *
#> 8 3 ( 2 ) 0.6423 0.6173 7.727757 11.01 * * *
#> 9 3 ( 3 ) 0.6191 0.5925 7.973957 14.25 * * *
#> 10 4 ( 1 ) 0.6993 0.6707 7.168166 5.03 * * * *
#> 11 4 ( 2 ) 0.6639 0.6319 7.579356 9.99 * * * *
#> 12 4 ( 3 ) 0.6498 0.6164 7.736422 11.96 * * * *
#> 13 5 ( 1 ) 0.7067 0.6710 7.165369 6.00 * * * * *
#
mdl <- lm(Sepal.Length ~ ., data = iris)
sumry(mdl)
#>
#> Summary Statistics:
#> Value Performance Measure Err(Resids) Metric
#> Observations = 150 R-Squared = 0.86731 MAPE = 0.041785
#> F-Statistic = 188.25 Adj-R2 = 0.86271 MAD = 0.24286
#> Pr(b's=0) = <2e-16 *** Std.Err.Est = 0.30683 RMSE = 0.30063
#>
#> Analysis of Variance:
#> Deg.Frdm Sum.of.Sqs Mean.Sum.Sqs F.statistic p-value(F)
#> Regression 5 88.612 17.722370 188.25 <2e-16 ***
#> Error(Resids) 144 13.556 0.094142
#> Total 149 102.168
#>
#> Coefficients:
#> Coefficient Std.Error t-stat p-value VIF
#> (Intercept) 2.17127 0.279794 7.7602 1.43e-12 ***
#> Sepal.Width 0.49589 0.086070 5.7615 4.87e-08 *** 2.2275
#> Petal.Length 0.82924 0.068528 12.1009 < 2e-16 *** 23.1616
#> Petal.Width -0.31516 0.151196 -2.0844 0.03889 * 21.0214
#> Species_versicolor -0.72356 0.240169 -3.0127 0.00306 ** 20.4234
#> Species_virginica -1.02350 0.333726 -3.0669 0.00258 ** 39.4344
#>
#> Signif.Levels: 0 '***' 0.001 '** ' 0.01 ' * ' 0.05 ' . ' 0.1 ' ' 1
#>
#> Summary of Min 1Q Mean Median 3Q Max
#> Residuals: -0.7942 -0.2187 <3e-14 0.008987 0.2025 0.731
#>
#> Call: lm(formula = Sepal.Length ~ ., data = iris)
#
mdl <- lm(mpg ~ hp + qsec, data = mtcars)
sumry(mdl)
#>
#> Summary Statistics:
#> Value Performance Measure Err(Resids) Metric
#> Observations = 32 R-Squared = 0.63688 MAPE = 0.1448
#> F-Statistic = 25.431 Adj-R2 = 0.61183 MAD = 2.6984
#> Pr(b's=0) = 4.18e-07 *** Std.Err.Est = 3.755 RMSE = 3.5746
#>
#> Analysis of Variance:
#> Deg.Frdm Sum.of.Sqs Mean.Sum.Sqs F.statistic p-value(F)
#> Regression 2 717.15 358.58 25.431 4.18e-07 ***
#> Error(Resids) 29 408.89 14.10
#> Total 31 1126.05
#>
#> Coefficients:
#> Coefficient Std.Error t-stat p-value VIF
#> (Intercept) 48.323705 11.103306 4.3522 0.000153 ***
#> hp -0.084593 0.013933 -6.0715 1.31e-06 *** 2.0063
#> qsec -0.886580 0.534585 -1.6584 0.108007 2.0063
#>
#> Signif.Levels: 0 '***' 0.001 '** ' 0.01 ' * ' 0.05 ' . ' 0.1 ' ' 1
#>
#> Summary of Min 1Q Median Mean 3Q Max
#> Residuals: -5.178 -2.603 -0.5098 <6e-15 1.287 8.718
#>
#> Call: lm(formula = mpg ~ hp + qsec, data = mtcars)
lm_plot.4way(mdl)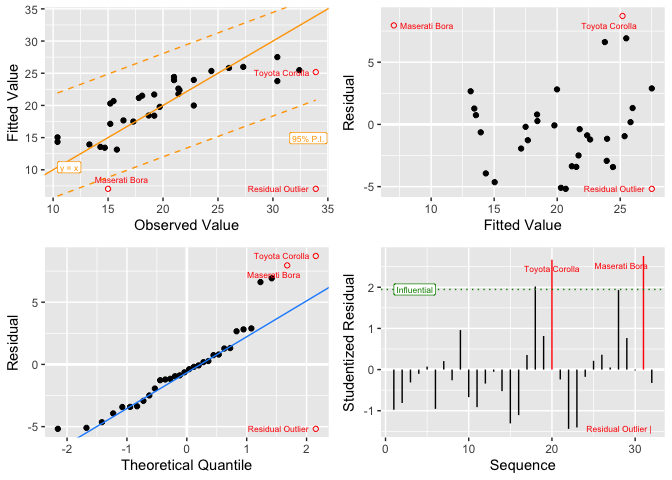
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.