The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
Access, download and locally cache files deposited on Zenodo easily.
You can install the development version of zendown from GitHub with:
# install.packages("remotes")
remotes::install_github("rfsaldanha/zendown")library(zendown)This Zenodo deposit contains RDS files from examples datasets.
First, you need to find the Zenodo deposit code. It is the number that appears on the end of a Zenodo deposit link. The code number also appears on the Zenodo DOI.
https://zenodo.org/records/10959197Deposition code: 10959197
With the deposit code and the desired file name, you can just access
the file with the zen_file function.
my_iris <- zen_file(deposit_id = 10959197, file_name = "iris.rds") |>
readRDS()
head(my_iris)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3.0 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 5.0 3.6 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosaThe function will create a cache on your machine with all accessed files, avoiding re-downloading them when you access some file again.
By default, the cache is stored on a temporary folder that is cleaned when the R session is ended.
To use a persistent cache and other options, check this article.
# https://zenodo.org/records/10848
zen_file(10848, "DOAJ_Soc_Licenses_Correct.csv") |>
read.csv2() |>
tibble::tibble() |>
head()
#> # A tibble: 6 × 12
#> X DOAJNR Title Title.Alternative Identifier Publisher Language ISSN EISSN
#> <int> <int> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 1 37 "Est… <NA> http://ww… "Univers… Spanish… 1405… <NA>
#> 2 2 65 "Rev… "Review of Resea… http://rc… "Expert … English… 1583… 1584…
#> 3 3 85 "Soc… <NA> http://ej… "Firenze… English… 2038… <NA>
#> 4 4 106 "Jou… <NA> http://jf… "New Pra… English 1945… 1944…
#> 5 5 195 "Oñ… <NA> http://op… "Oñati … English… 2079… <NA>
#> 6 6 198 "Soc… <NA> http://so… "Associa… French 1950… <NA>
#> # ℹ 3 more variables: CC.License.Doaj <chr>, CC.License.Checked <chr>,
#> # Wrong.Cat.Doaj <chr># https://zenodo.org/records/10947952
zen_file(10947952, "2m_temperature_max.parquet") |>
arrow::read_parquet() |>
tibble::tibble() |>
head()
#> # A tibble: 6 × 4
#> code_muni date name value
#> <int> <date> <chr> <dbl>
#> 1 1100015 2023-01-01 2m_temperature_max_mean 304.
#> 2 1100015 2023-01-02 2m_temperature_max_mean 302.
#> 3 1100015 2023-01-03 2m_temperature_max_mean 302.
#> 4 1100015 2023-01-04 2m_temperature_max_mean 302.
#> 5 1100015 2023-01-05 2m_temperature_max_mean 297.
#> 6 1100015 2023-01-06 2m_temperature_max_mean 302.# https://zenodo.org/records/10889682
zen_file(10889682, "total_precipitation_2023-09-01_2023-09-30_day_sum.nc") |>
terra::rast() |>
terra::plot(1)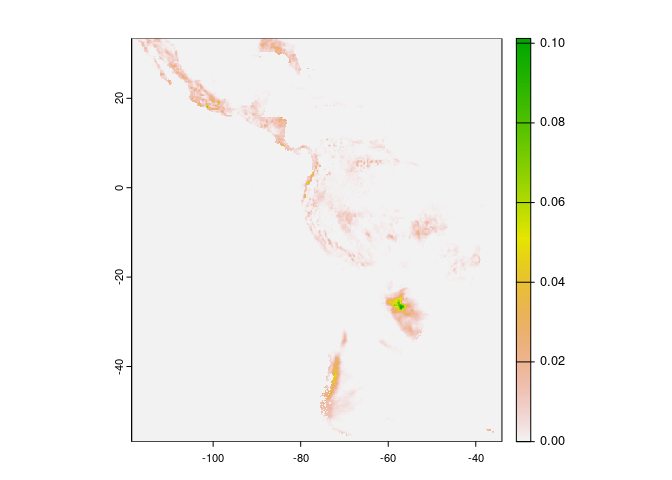
To explore the Zenodo API possibilities to problematically store files and other procedures, check the zen4R package.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.