The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
wsMed
The wsMed function is designed for two condition
within-subject mediation analysis, incorporating SEM models through the
lavaan package and Monte Carlo simulation methods. This
document provides a detailed description of the function’s parameters,
workflow, and usage, along with an example demonstration.
Installation
You can install the development version of wsMed from GitHub with:
# install.packages("pak")
pak::pak("Yangzhen1999/wsMed")
Alternatively, if you prefer using devtools, you can install wsMed as
follows:
# install.packages("devtools")
devtools::install_github("Yangzhen1999/wsMed")
Example
This is a basic example which shows you how to solve a common
problem:
library(wsMed)
# Load example data
data(example_data)
set.seed(123)
example_dataN <- mice::ampute(
data = example_data,
prop = 0.1,
)$amp
# Perform within-subject mediation analysis (Parallel mediation model)
result <- wsMed(
data = example_dataN, #dataset
M_C1 = c("A1","B1"), # A1/B1 is A/B mediator variable in condition 1
M_C2 = c("A2","B2"), # A2/B2 is A/B mediator variable in condition 2
Y_C1 = "C1", # C1 is outcome variable in condition 1
Y_C2 = "C2", # C2 is outcome variable in condition 2
form = "P", # Parallel mediation
C_C1 = "D1", # within-subject covariate (e.g., measured under D1)
C_C2 = "D2", # within-subject covariate (e.g., measured under C2)
C = "D3", # between-subject covariates
Na = "MI", # Use multiple imputation for missing data
standardized = TRUE, # Request standardized path coefficients and effects
)
# Print summary results
print(result)
Main Function Overview
The wsMed() function automates the full workflow for
two-condition within-subject mediation analysis. Its main steps are:
Validate inputs – check dataset structure,
mediation model type (form), and missing-data
settings.
Prepare data – compute difference scores
(Mdiff, Ydiff) and centered averages
(Mavg) from the two-condition variables.
Build the model – generate SEM syntax according
to the chosen structure:
"P": Parallel mediation
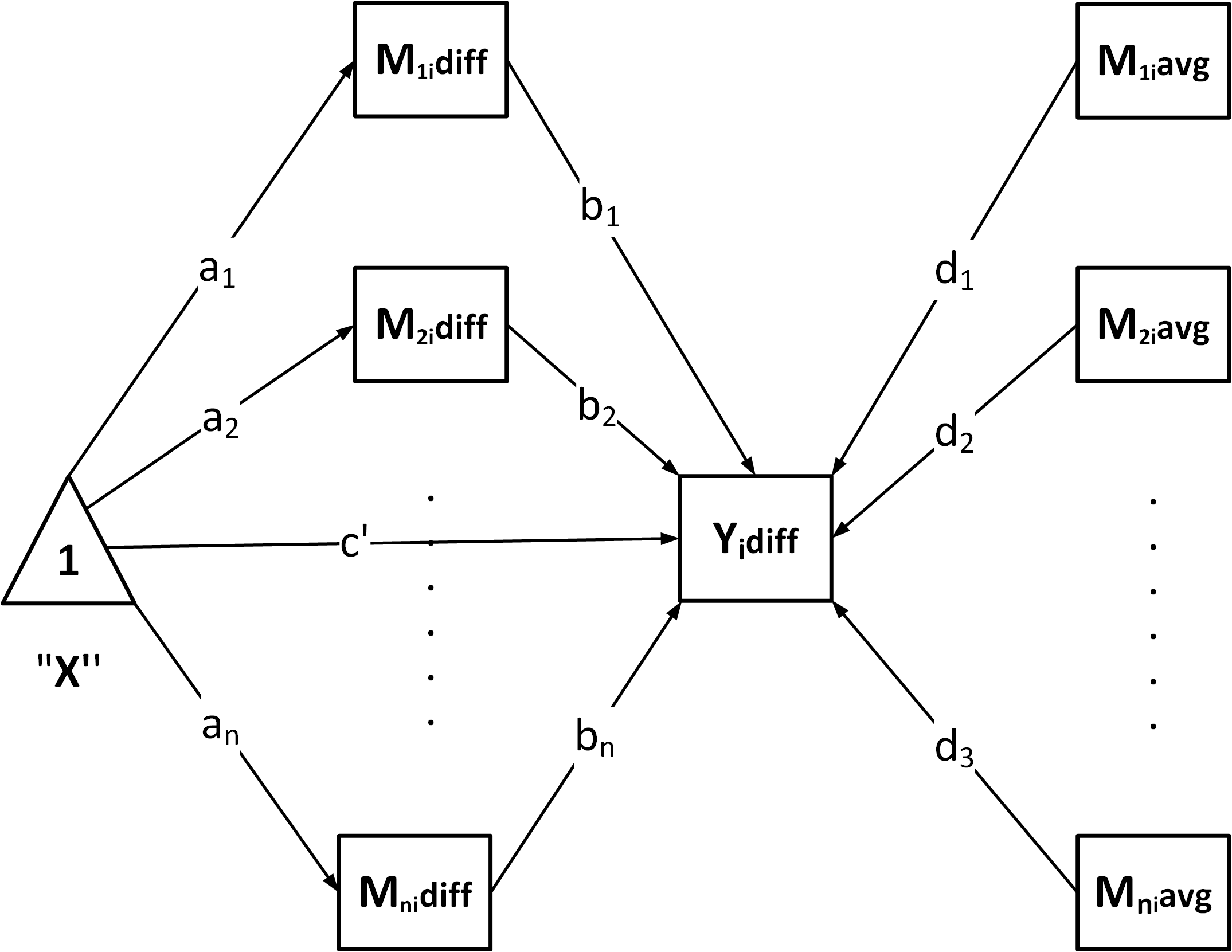
"CN": Chained / serial mediation
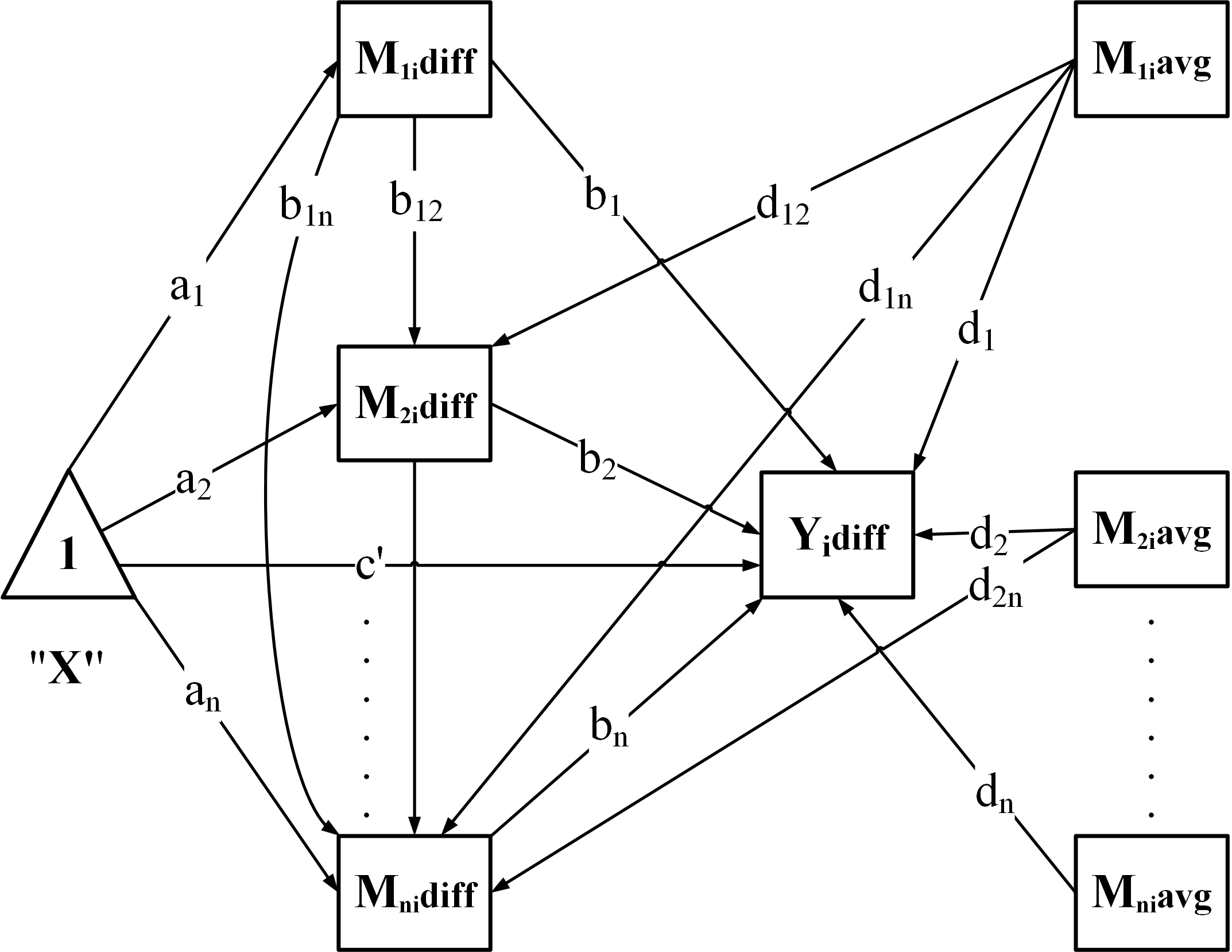
"CP": Chained + Parallel
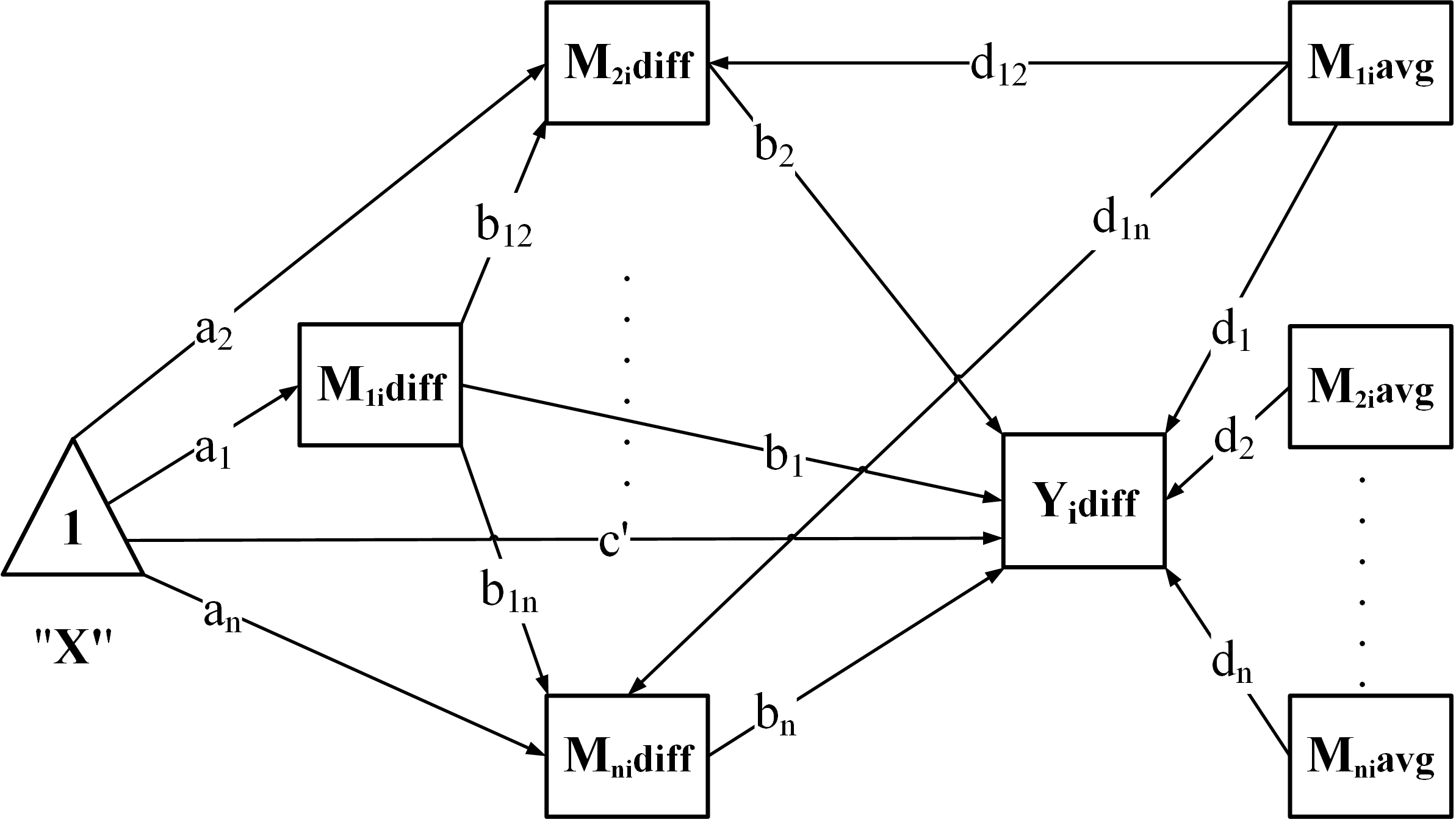
"PC": Parallel + Chained
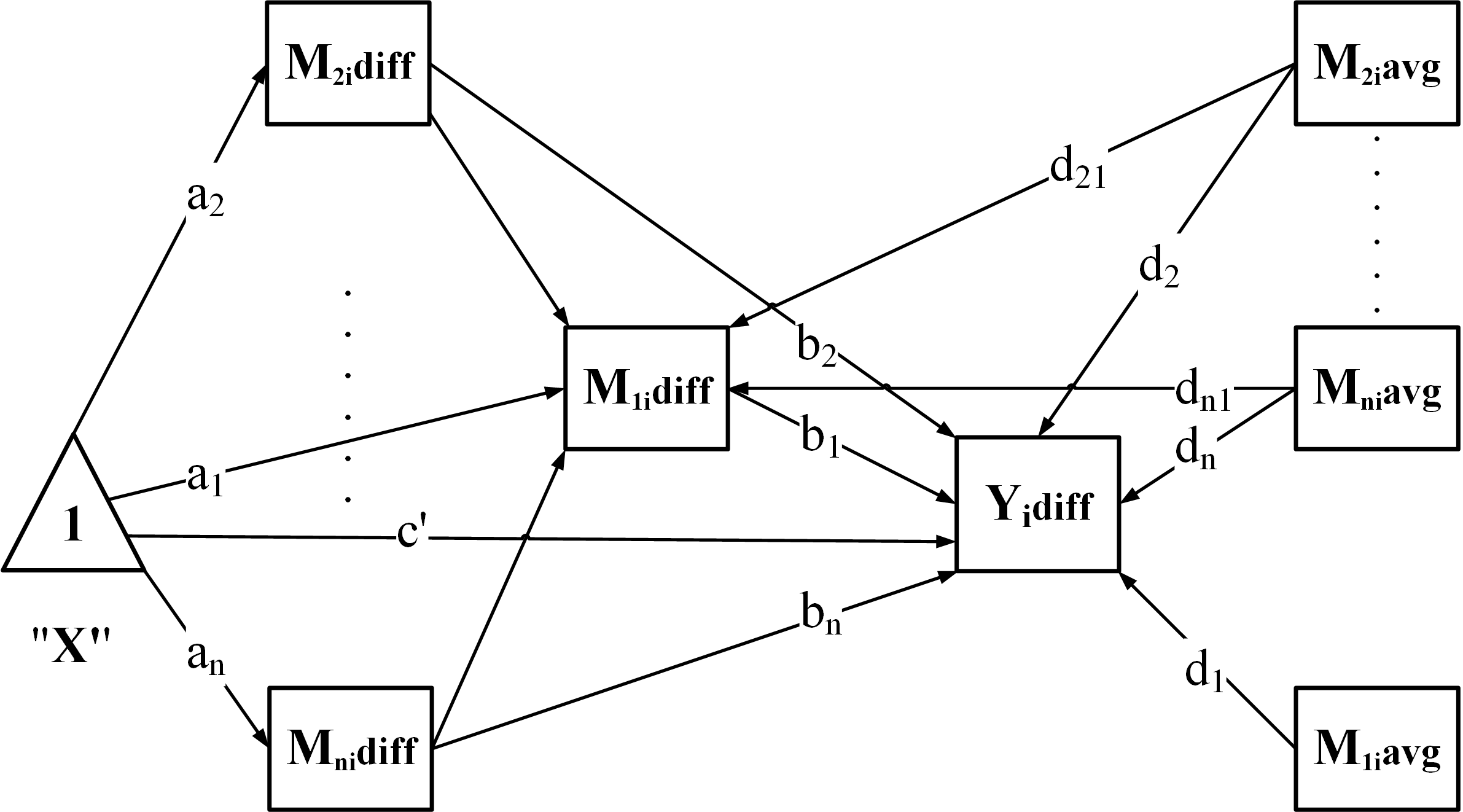
Fit the model – estimate parameters while
handling missing data:
"DE": listwise deletion"FIML": full-information ML"MI": multiple imputation
Compute inference – provide confidence intervals
using:
- Bootstrap
(
ci_method = "bootstrap")
- Monte Carlo (
ci_method = "mc")
Optional: Standardization – if
standardized = TRUE, return standardized effects with
CIs.
Optional: Covariates – automatically center and
include:
- Between-subject covariates (
C):
mean-centered and added to all regressions.
- Within-subject covariates (
C_C1,
C_C2): difference scores and centered averages are computed
and included.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.



