The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
This package allows users to create, visualize, and evaluate regression trees and random forests for longitudinal or functional data through a spline projection method first suggested by Yu and Lambert (1999).
You can install splinetree from CRAN or from github with:
# install.packages("devtools")
devtools::install_github("anna-neufeld/splinetree")Detailed information on using this package can be found in the package vignettes. The package vignettes can be accessed with:
browseVignettes(package='splinetree')The vignettes are also available on the package website, https://anna-neufeld.github.io/splinetree/reference/index.html.
library(splinetree)
#> Loading required package: rpart
#> Loading required package: nlme
#> Loading required package: splines
tree1 <- splineTree(~HISP+WHITE+BLACK+HGC_MOTHER+HGC_FATHER+SEX+Num_sibs,
BMI ~ AGE, "ID", nlsySample, degree = 1, df=2, intercept = FALSE, cp = 0.005)
stPrint(tree1)
#> n= 1000,
#>
#> node), split, n , coefficients
#> * denotes terminal node
#>
#> 1) root, 1000, (4.961796, 8.091247)
#> 2) WHITE< 0.5, 505, (5.882807, 9.006190)*
#> 3) WHITE>=0.5, 495, (4.022179, 7.157821)
#> 6) HGC_FATHER< 8.5, 78, (5.198284, 8.642817)*
#> 7) HGC_FATHER>=8.5, 417, (3.802188, 6.880053)*stPlot(tree)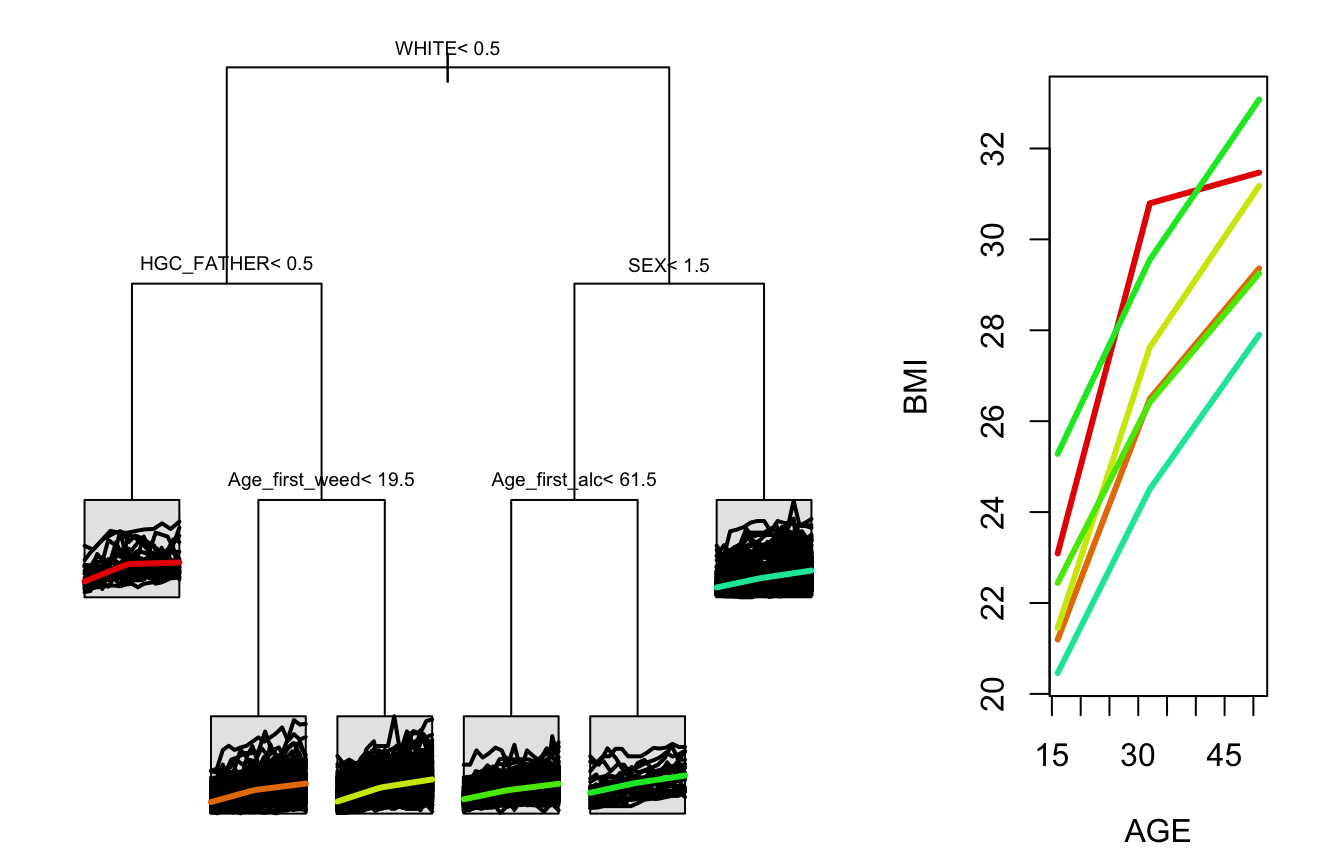
set.seed(1234)
forest1 <- splineForest(~HISP+WHITE+BLACK+HGC_MOTHER+HGC_FATHER+SEX+Num_sibs,
BMI ~ AGE, "ID", nlsySample, degree = 1, df=2, intercept = FALSE, ntree=50, prob=1/2)
varImps <- varImpCoeff(forest1)plotImp(varImps[,3])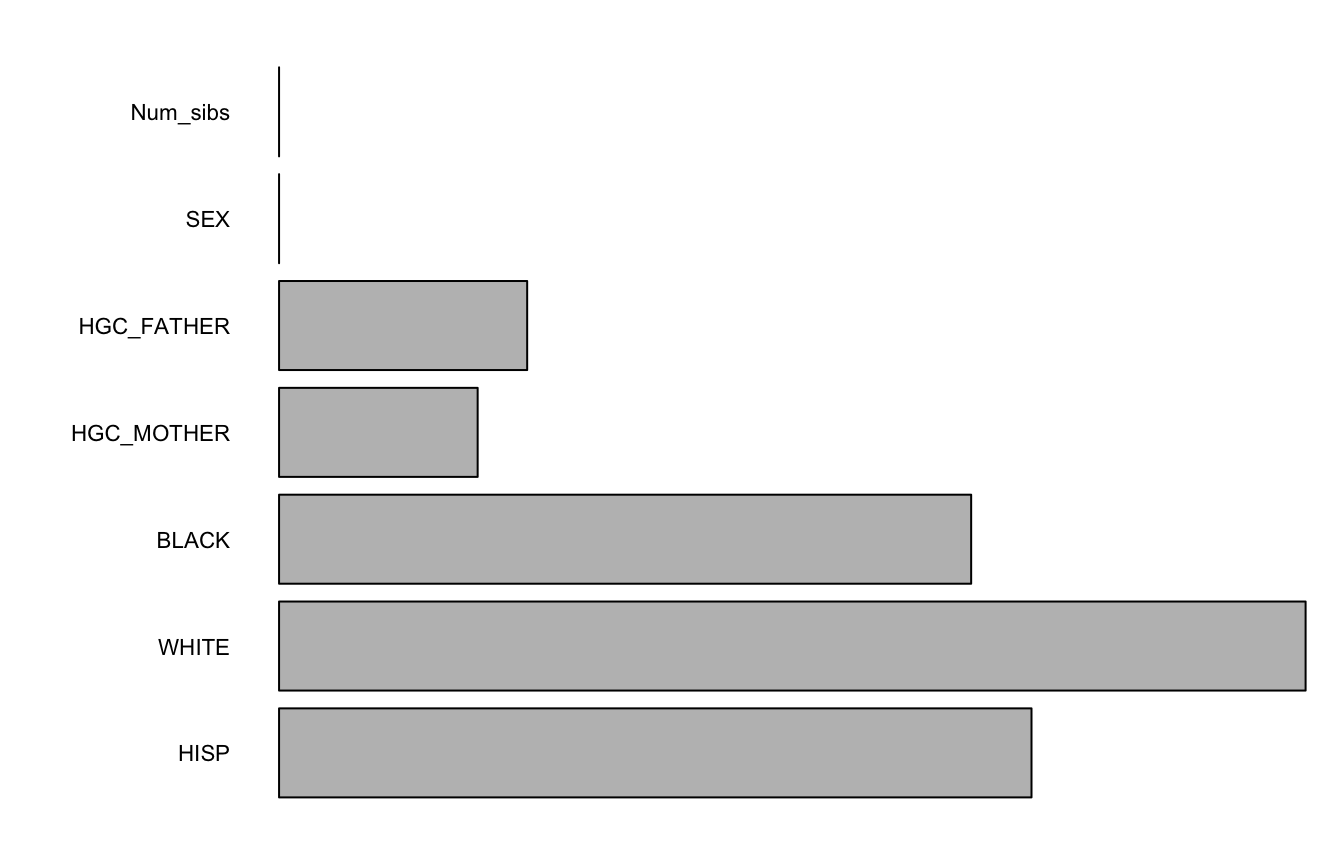
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.