The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
Repo is a data-centered data flow manager. It allows to store R data files in a central local repository, together with tags, annotations, provenance and dependence information. Any saved object can then be easily located and loaded through the repo interface.
A paper about Repo has been published in BMC Bioinformatics.
Latest news are found in the NEWS.md file of the “Untested” branch.
Creating a dummy repository under the R temporary folder (skipping confirmation):
library(repo)
rp <- repo_open(tempdir(), force=T)
#> Repo created.Storing data. In this case, just item values and names are specified:
God <- Inf
rp$put(God) ## item name inferred from variable name
rp$put(0, "user") ## item name specifiedMore data with specified dependencies:
rp$put(pi, "The Pi costant", depends="God")
rp$put(1:10, "r", depends="user")Loading items from the repository on the fly using names:
diam <- 2 * rp$get("r")
circum <- 2 * rp$get("The Pi costant") * rp$get("r")
area <- rp$get("The Pi costant") * rp$get("r") ^ 2Storing more data with verbose descriptions:
rp$put(diam, "diameters", "These are the diameters", depends = "r")
rp$put(circum, "circumferences", "These are the circumferences",
depends = c("The Pi costant", "r"))
rp$put(area, "areas", "These are the areas",
depends = c("The Pi costant", "r"))Showing repository contents:
print(rp)
#> ID Dims Size
#> God 1 51 B
#> user 1 49 B
#> The Pi costant 1 55 B
#> r 10 99 B
#> diameters 10 75 B
#> circumferences 10 103 B
#> areas 10 103 Brp$info()
#> Root: /tmp/RtmppFMwnb
#> Number of items: 7
#> Total size: 535 Brp$info("areas")
#> ID: areas
#> Description: These are the areas
#> Tags:
#> Dimensions: 10
#> Timestamp: 2019-12-22 17:01:45
#> Size on disk: 103 B
#> Provenance:
#> Attached to: -
#> Stored in: /tmp/RtmppFMwnb/a/areas
#> MD5 checksum: 56ad410055fedb0cae012d813a130291
#> URL: -Visualizing dependencies:
rp$dependencies()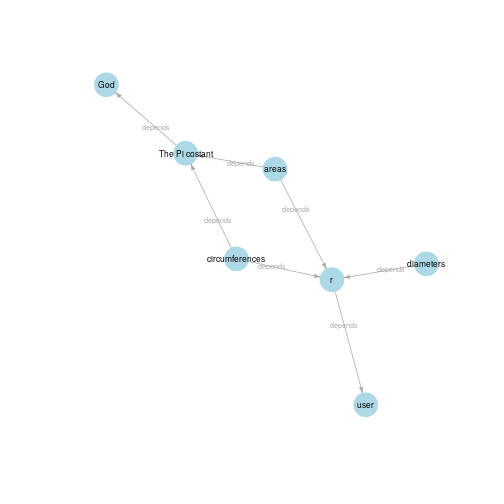
Manual acces to stored data:
fpath <- rp$attr("r", "path")
readRDS(fpath)
#> [1] 1 2 3 4 5 6 7 8 9 10Master: stable major releases, usually in sync with the latest CRAN version.
Dev: minor releases passing automatic checks.
Untested: in progress versions and prototype code, not necessarily working.
Besides inline help, two documents are available as introductory material:
Repo is on CRAN and can be installed from within R as follows:
install.packages("repo")Latest stable release can be downloaded from Github at https://github.com/franapoli/repo. Repo can then be installed from the downloaded sources as follows:
install.packages("path-to-downloaded-source", repos=NULL)devtools users can install Repo directly from github as
follows:
install_github("franapoli/repo", ref="dev")These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.