The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
A data package which lists every command in base R packages since R version 0.50.
The latest R version covered is 4.4.0.
You can view the data online in a Shiny app.
From r-universe:
install.packages('rcheology', repos = c('https://hughjonesd.r-universe.dev',
'https://cloud.r-project.org'))From CRAN:
R versions are built using the evercran project.
Results are found from running ls(all.names = TRUE) on all installed packages. For more details, see guest-list-objects.R.
The Rversions data frame lists versions of R and release dates.
host-run-on-evercran.sh.library(rcheology)
data("rcheology")
rcheology[rcheology$name == "kmeans" & rcheology$Rversion %in% c("1.0.1", "1.9.0", "2.1.0", "3.0.2", "3.2.0", "4.0.2"), ]
#> package name Rversion priority type exported hidden class
#> 338100 mva kmeans 1.0.1 <NA> closure TRUE FALSE <NA>
#> 388198 stats kmeans 1.9.0 base closure TRUE FALSE function
#> 388202 stats kmeans 2.1.0 base closure TRUE FALSE function
#> 388242 stats kmeans 3.0.2 base closure TRUE FALSE function
#> 388248 stats kmeans 3.2.0 base closure TRUE FALSE function
#> 388273 stats kmeans 4.0.2 base closure TRUE FALSE function
#> S4generic
#> 338100 FALSE
#> 388198 FALSE
#> 388202 FALSE
#> 388242 FALSE
#> 388248 FALSE
#> 388273 FALSE
#> args
#> 338100 (x, centers, iter.max = 10)
#> 388198 (x, centers, iter.max = 10)
#> 388202 (x, centers, iter.max = 10, nstart = 1, algorithm = c("Hartigan-Wong", "Lloyd", "Forgy", "MacQueen"))
#> 388242 (x, centers, iter.max = 10, nstart = 1, algorithm = c("Hartigan-Wong", "Lloyd", "Forgy", "MacQueen"), trace = FALSE)
#> 388248 (x, centers, iter.max = 10L, nstart = 1L, algorithm = c("Hartigan-Wong", "Lloyd", "Forgy", "MacQueen"), trace = FALSE)
#> 388273 (x, centers, iter.max = 10L, nstart = 1L, algorithm = c("Hartigan-Wong", "Lloyd", "Forgy", "MacQueen"), trace = FALSE)Latest changes:
suppressPackageStartupMessages(library(dplyr))
r_penultimate <- sort(package_version(unique(rcheology::rcheology$Rversion)),
decreasing = TRUE)
r_penultimate <- r_penultimate[2]
r_latest_obj <- rcheology %>% dplyr::filter(Rversion == r_latest)
r_penult_obj <- rcheology %>% dplyr::filter(Rversion == r_penultimate)
r_introduced <- anti_join(r_latest_obj, r_penult_obj, by = c("package", "name"))
r_introduced
#> # A tibble: 19 × 10
#> package name Rversion priority type exported hidden class S4generic args
#> <chr> <chr> <chr> <chr> <chr> <lgl> <lgl> <chr> <lgl> <chr>
#> 1 Matrix .__C_… 4.4.0 recomme… S4 TRUE TRUE clas… FALSE <NA>
#> 2 base %||% 4.4.0 base clos… TRUE FALSE func… FALSE "(x,…
#> 3 base .inte… 4.4.0 base char… TRUE TRUE char… FALSE <NA>
#> 4 base .rang… 4.4.0 base clos… TRUE TRUE func… FALSE "(..…
#> 5 base Exec 4.4.0 base spec… TRUE FALSE func… NA "(ex…
#> 6 base Tailc… 4.4.0 base spec… TRUE FALSE func… NA "(FU…
#> 7 base decla… 4.4.0 base spec… TRUE FALSE func… NA "(..…
#> 8 base range… 4.4.0 base clos… TRUE FALSE func… FALSE "(..…
#> 9 base range… 4.4.0 base clos… TRUE FALSE func… FALSE "(..…
#> 10 base sort_… 4.4.0 base clos… TRUE FALSE func… FALSE "(x,…
#> 11 base sort_… 4.4.0 base clos… TRUE FALSE func… FALSE "(x,…
#> 12 base sort_… 4.4.0 base clos… TRUE FALSE func… FALSE "(x,…
#> 13 base use 4.4.0 base clos… TRUE FALSE func… FALSE "(pa…
#> 14 parallel close… 4.4.0 base clos… TRUE FALSE func… FALSE "(no…
#> 15 parallel recvD… 4.4.0 base clos… TRUE FALSE func… FALSE "(no…
#> 16 parallel recvO… 4.4.0 base clos… TRUE FALSE func… FALSE "(cl…
#> 17 parallel sendD… 4.4.0 base clos… TRUE FALSE func… FALSE "(no…
#> 18 tools pkg2H… 4.4.0 base clos… TRUE FALSE func… FALSE "(pa…
#> 19 tools stand… 4.4.0 base clos… TRUE FALSE func… FALSE "()"Base functions over time:
library(ggplot2)
rvs <- rcheology$Rversion %>%
unique() %>%
as.package_version() %>%
sort() %>%
as.character()
major_rvs <- grep(".0$", rvs, value = TRUE)
major_rvs <- gsub("0.50", "0.50-a1", major_rvs)
major_rvs <- gsub("0.60", "0.60.0", major_rvs)
major_rv_dates <- Rversions$date[Rversions$Rversion %in% major_rvs]
major_rv_dates
#> [1] "1997-07-22" "1997-12-04" "1997-12-22" "1998-11-14" "1999-04-08"
#> [6] "1999-08-28" "1999-11-22" "2000-02-07" "2000-02-29" "2000-06-15"
#> [11] "2000-12-15" "2001-06-22" "2001-12-19" "2002-04-29" "2002-10-01"
#> [16] "2003-04-16" "2003-10-08" "2004-04-12" "2004-10-04" "2005-04-19"
#> [21] "2005-10-06" "2006-04-24" "2006-10-03" "2007-04-24" "2007-10-03"
#> [26] "2008-04-22" "2008-10-20" "2009-04-17" "2009-10-26" "2010-04-22"
#> [31] "2010-10-15" "2011-04-13" "2011-10-31" "2012-03-30" "2013-04-03"
#> [36] "2014-04-10" "2015-04-16" "2016-05-03" "2017-04-21" "2018-04-23"
#> [41] "2019-04-26" "2020-04-24" "2021-05-18" "2022-04-22" "2023-04-21"
#> [46] "2024-04-24"
major_rvs <- gsub("\\.0$", "", major_rvs)
rch_dates <- rcheology %>% left_join(Rversions, by = "Rversion")
ggplot(rch_dates, aes(date, group = package, fill = package), colour = NA) +
stat_count(geom = "area") +
theme(axis.text.x = element_text(angle = 45, hjust = 1, size = 8)) +
# ggthemes::scale_fill_gdocs() +
scale_x_date(breaks = major_rv_dates, labels = major_rvs) +
xlab("Version") + ylab("Function count") +
theme(legend.position = "top")
#> Warning: Removed 1615 rows containing non-finite outside the scale range
#> (`stat_count()`).
An alternative view:
ggplot(rch_dates, aes(date, fill = "orange")) +
stat_count(geom = "area") +
scale_x_date(breaks = major_rv_dates, labels = major_rvs) +
theme(axis.text.x = element_text(angle = 45, hjust = 1, size = 8)) +
xlab("Version") + ylab("Function count") +
facet_wrap(~package, scales = "free_y", ncol = 2) +
theme(legend.position = "none")
#> Warning: Removed 1615 rows containing non-finite outside the scale range
#> (`stat_count()`).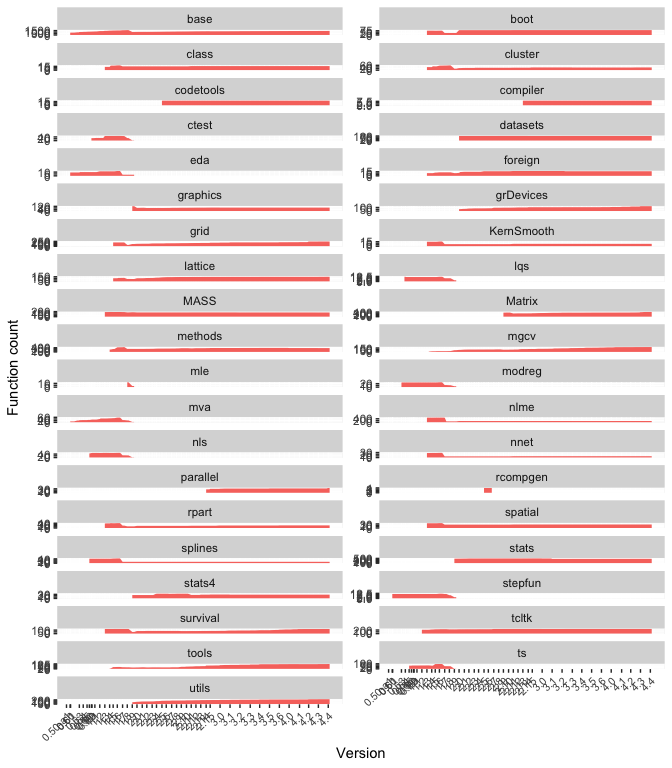
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.