The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
This R package produces nice looking parameter tables for pharmacometric modeling results with ease. It is completely agnostic to the modeling software that was used.
This package is not yet on CRAN. To install the latest development version directly from GitHub:
require(remotes)
remotes::install_github("benjaminrich/pmxpartab")The creation of the parameter table proceeds in 2 steps:
data.frame from model outputs
and metadata.data.frame.Both the model outputs and metadata are provided as R lists (it is a separate problem to extract the outputs from the modeling software into the required list format).
Here is an example (in this example, YAML is used to give a clear and concise representation of outputs and metadata, but this is not required).
library(yaml)
outputs <- yaml.load("
est:
CL: 0.482334
VC: 0.0592686
CL_WT: 0.750000
VC_WT: 1.00000
nCL: 0.315414
nVC: 0.536025
ERRP: 0.0508497
se:
CL: 0.0138646
VC: 0.0055512
nCL: 0.0188891
nVC: 0.0900352
ERRP: 0.0018285
fixed:
CL: no
VC: no
CL_WT: yes
VC_WT: yes
nCL: no
nVC: no
ERRP: no
shrinkage:
nCL: 9.54556
nVC: 47.8771
")
meta <- yaml.load("
parameters:
- name: CL
label: 'Clearance'
units: 'L/h'
type: Structural
- name: VC
label: 'Volume'
units: 'L'
type: Structural
trans: 'exp'
- name: CL_WT
label: 'Weight on Clearance'
type: CovariateEffect
- name: VC_WT
label: 'Weight on Volume'
type: CovariateEffect
- name: nCL
label: 'On Clearance'
type: IIV
trans: 'SD (CV%)'
- name: nVC
label: 'On Volume'
type: IIV
trans: 'SD (CV%)'
- name: ERRP
label: 'Proportional Error'
units: '%'
type: RUV
trans: '%'
")
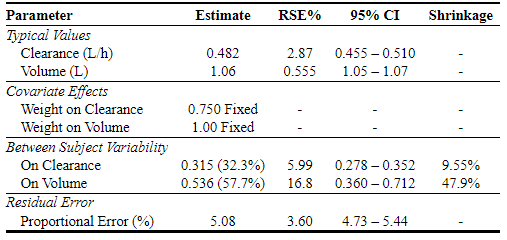
parframe <- pmxparframe(outputs, meta)
parframe
#> name label units type trans fixed est
#> 1 CL Clearance L/h Structural <NA> FALSE 0.482334
#> 2 VC Volume L Structural exp FALSE 1.061060
#> 3 CL_WT Weight on Clearance <NA> CovariateEffect <NA> TRUE 0.750000
#> 4 VC_WT Weight on Volume <NA> CovariateEffect <NA> TRUE 1.000000
#> 5 nCL On Clearance <NA> IIV SD (CV%) FALSE 0.315414
#> 6 nVC On Volume <NA> IIV SD (CV%) FALSE 0.536025
#> 7 ERRP Proportional Error % RUV % FALSE 5.084970
#> se rse lci95 uci95 pval shrinkage
#> 1 0.013864600 2.874481 0.4551594 0.5095086 0.000000e+00 NA
#> 2 0.005890157 0.555120 1.0495781 1.0726679 0.000000e+00 NA
#> 3 NA NA NA NA NA NA
#> 4 NA NA NA NA NA NA
#> 5 0.018889100 5.988669 0.2783914 0.3524366 0.000000e+00 9.54556
#> 6 0.090035200 16.796829 0.3595560 0.7124940 2.624601e-09 47.87710
#> 7 0.182850000 3.595891 4.7265840 5.4433560 0.000000e+00 NApmxpartab(parframe)Which produces:

For more information, read the vignette.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.