The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

The ‘personalized’ package is designed for the analysis of data where the effect of a treatment or intervention may vary for different patients. It can be used for either data from randomized controlled trials or observational studies and is not limited specifically to the analysis of medical data.
The personalized package provides estimation methods for subgroup identification under the framework of Chen et al (2017). It also provides routines for valid estimation of the subgroup-specific treatment effects.
Tutorial / Vignette - tutorial of statistical methodology and usage of the package
Install from CRAN using:
install.packages("personalized")or install the development version using the devtools package:
devtools::install_github("jaredhuling/personalized")or by cloning and building using R CMD INSTALL
Load the package:
library(personalized)(it should be a function which inputs covariates and treatments and returns propensity score):
prop.func <- function(x, trt)
{
# fit propensity score model
propens.model <- cv.glmnet(y = trt,
x = x, family = "binomial")
pi.x <- predict(propens.model, s = "lambda.min",
newx = x, type = "response")[,1]
pi.x
}subgrp.model <- fit.subgroup(x = x, y = y,
trt = trt,
propensity.func = prop.func,
loss = "sq_loss_lasso",
nfolds = 5) # option for cv.glmnetsummary(subgrp.model)## family: gaussian
## loss: sq_loss_lasso
## method: weighting
## cutpoint: 0
## propensity
## function: propensity.func
##
## benefit score: f(x),
## Trt recom = Trt*I(f(x)>c)+Ctrl*I(f(x)<=c) where c is 'cutpoint'
##
## Average Outcomes:
## Recommended Ctrl Recommended Trt
## Received Ctrl -3.9319 (n = 109) -21.2055 (n = 122)
## Received Trt -25.078 (n = 112) -8.326 (n = 157)
##
## Treatment effects conditional on subgroups:
## Est of E[Y|T=Ctrl,Recom=Ctrl]-E[Y|T=/=Ctrl,Recom=Ctrl]
## 21.1461 (n = 221)
## Est of E[Y|T=Trt,Recom=Trt]-E[Y|T=/=Trt,Recom=Trt]
## 12.8795 (n = 279)
##
## NOTE: The above average outcomes are biased estimates of
## the expected outcomes conditional on subgroups.
## Use 'validate.subgroup()' to obtain unbiased estimates.
##
## ---------------------------------------------------
##
## Benefit score quantiles (f(X) for Trt vs Ctrl):
## 0% 25% 50% 75% 100%
## -9.2792 -1.8237 0.5011 2.5977 9.6376
##
## ---------------------------------------------------
##
## Summary of individual treatment effects:
## E[Y|T=Trt, X] - E[Y|T=Ctrl, X]
##
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -18.5583 -3.6474 1.0023 0.9507 5.1954 19.2753
##
## ---------------------------------------------------
##
## 5 out of 50 interactions selected in total by the lasso (cross validation criterion).
##
## The first estimate is the treatment main effect, which is always selected.
## Any other variables selected represent treatment-covariate interactions.
##
## Trt V2 V11 V17 V32 V35
## Estimate 0.5463 0.9827 -0.4356 -0.1532 0.0326 0.1007val.model <- validate.subgroup(subgrp.model, B = 100,
method = "training_test",
train.fraction = 0.75)print(val.model, digits = 2, sample.pct = TRUE)## family: gaussian
## loss: sq_loss_lasso
## method: weighting
##
## validation method: training_test_replication
## cutpoint: 0
## replications: 100
##
## benefit score: f(x),
## Trt recom = Trt*I(f(x)>c)+Ctrl*I(f(x)<=c) where c is 'cutpoint'
##
## Average Test Set Outcomes:
## Recommended Ctrl Recommended Trt
## Received Ctrl -9.56 (SE = 7.98, 19.88%) -18.62 (SE = 6.72, 26.5%)
## Received Trt -16.64 (SE = 6.85, 23.23%) -13.41 (SE = 7.8, 30.39%)
##
## Treatment effects conditional on subgroups:
## Est of E[Y|T=Ctrl,Recom=Ctrl]-E[Y|T=/=Ctrl,Recom=Ctrl]
## 6.54 (SE = 10.49, 43.11%)
## Est of E[Y|T=Trt,Recom=Trt]-E[Y|T=/=Trt,Recom=Trt]
## 5.21 (SE = 11.06, 56.89%)
##
## Est of
## E[Y|Trt received = Trt recom] - E[Y|Trt received =/= Trt recom]:
## 2.91 (SE = 8.29)Visualize subgroup-specific treatment effect estimates across training/testing iterations:
plot(val.model)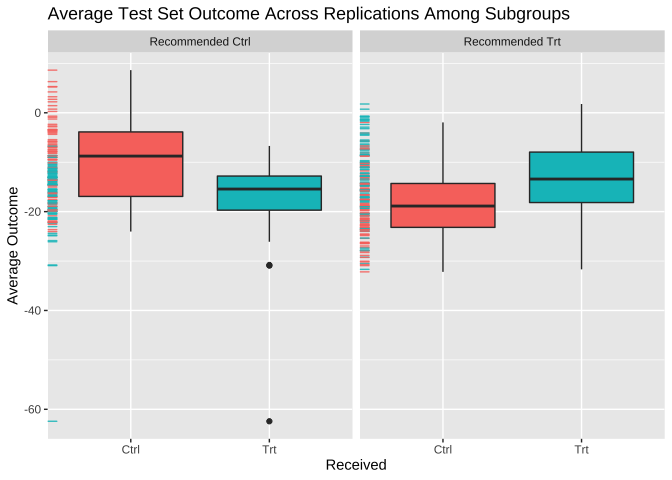
Here we only display covariates with a significantly different mean value (at level 0.05)
summ <- summarize.subgroups(subgrp.model)
print(summ, p.value = 0.05)## Avg (recom Ctrl) Avg (recom Trt) Ctrl - Trt SE (recom Ctrl) SE (recom Trt)
## V2 -2.4161 1.9013 -4.317 0.1423 0.1298
## V11 1.1279 -0.7963 1.924 0.1914 0.1572
## V17 0.8053 -0.3715 1.177 0.2170 0.1736personalizedAccess help files for the main functions of the
personalized package:
?fit.subgroup
?validate.subgroupThese binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.