
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

pctax provides a comprehensive suite of tools for
analyzing omics data.
The HTML documentation of the latest version is available at Github page.
Please go to https://bookdown.org/Asa12138/pctax_book/ for the full vignette.
The stable version is available on CRAN:
install.packages("pctax")Or you can install the development version of pctax from GitHub with:
# install.packages("devtools")
devtools::install_github("Asa12138/pctax")Recently, I added a function to draw two trees and their relationships:
data(otutab, package = "pcutils")
df2tree(taxonomy[1:50, ]) -> tax_tree
df2tree(taxonomy[51:100, ]) -> tax_tree2
link <- data.frame(from = sample(tax_tree$tip.label, 20), to = sample(tax_tree2$tip.label, 20))
plot_two_tree(tax_tree, tax_tree2, link,
tree1_tip = T, tree2_tip = T,
tip1_param = list(size = 2), tip2_param = list(size = 2)
)
Two trees and their relationships
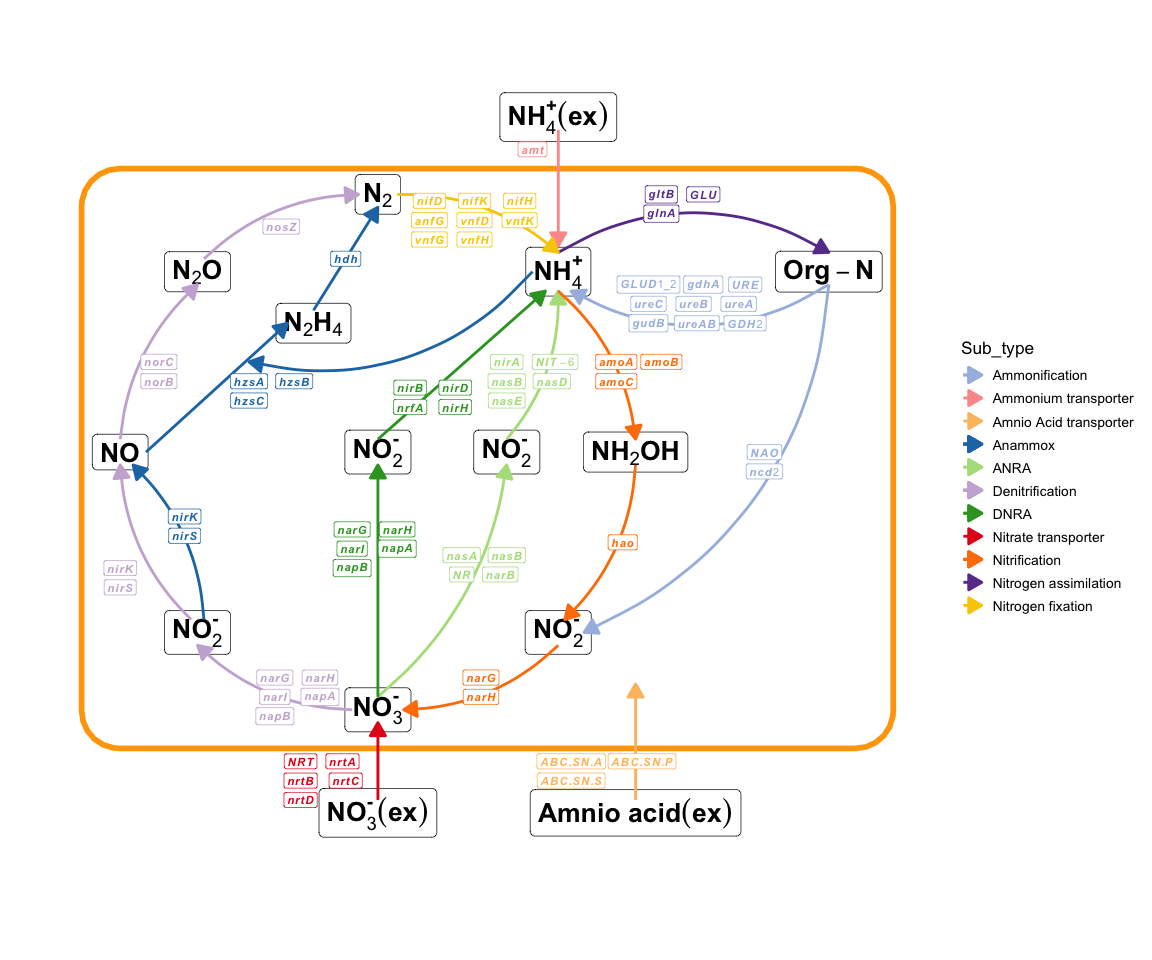
Recently I added a function to plot element cycling because element cycling genes are important in the microbiome (especially the environmental microbiome). Supports simple cycle diagram drawing of C, N, P, S, Fe (manual arrangement, there must be some missing parts, will be continuously added in the future):
plot_element_cycle(cycle = "Nitrogen cycle")
#> recommend ggsave(width = 12,height = 10)
Nitrogen cycle
For the full vignette, please visit pctax: Analyzing Omics Data with R.
Some Functionalities of pctax:

Please cite:
Chen Peng, Chao Jiang (2023). pctax: Professional Comprehensive Microbiome Data Analysis Pipeline. R package, https://github.com/Asa12138/pctax.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.