The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
This package acts as an interface to Our World in Data datasets, allowing for an easy way to search through data used in over 3,000 charts and load them into the R environment.
To install from CRAN:
install.packages("owidR")To install the development version from GitHub:
devtools::install_github("piersyork/owidR")The main function in owidR is owid(), which takes a
chart id and returns a data.table of the corresponding OWID dataset. To
search for chart ids you can use owid_search() to list all
the chart ids that match a keyword or regular expression.
Lets use the core functions to get data on how human rights have changed over time. First by searching for charts on human rights.
library(owidR)
owid_search("human rights")
## chart_id
## [1,] "human-rights-index-vs-electoral-democracy-index"
## [2,] "cases-of-killed-human-rights-defenders-journalists-trade-unionists"
## [3,] "countries-with-independent-national-human-rights-institution"
## [4,] "distribution-human-rights-index-vdem"
## [5,] "human-rights-index-vdem"
## [6,] "human-rights-index-population-weighted"
## [7,] "human-rights-index-vs-gdp-per-capita"
## [8,] "share-countries-accredited-independent-national-human-rights-institutions"
## title
## [1,] "Human rights index vs. electoral democracy index"
## [2,] "Confirmed killings of human rights defenders, journalists and trade unionists"
## [3,] "Countries with accredited independent national human rights institutions"
## [4,] "Distribution of human rights index"
## [5,] "Human rights index"
## [6,] "Human rights index"
## [7,] "Human rights index vs. GDP per capita"
## [8,] "Share of countries with accredited independent national human rights institutions"Let’s use the v-dem human rights index dataset.
rights <- owid("human-rights-index-vdem")
rights
## Key: <entity, code, year>
## entity code year civ_libs_vdem_owid civ_libs_vdem_high_owid
## <char> <char> <int> <num> <num>
## 1: Afghanistan AFG 1789 0.125 0.169
## 2: Afghanistan AFG 1790 0.125 0.169
## 3: Afghanistan AFG 1791 0.125 0.169
## 4: Afghanistan AFG 1792 0.125 0.169
## 5: Afghanistan AFG 1793 0.125 0.169
## ---
## 33373: Zimbabwe ZWE 2018 0.428 0.473
## 33374: Zimbabwe ZWE 2019 0.403 0.456
## 33375: Zimbabwe ZWE 2020 0.413 0.469
## 33376: Zimbabwe ZWE 2021 0.395 0.443
## 33377: Zimbabwe ZWE 2022 0.388 0.432
## civ_libs_vdem_low_owid
## <num>
## 1: 0.089
## 2: 0.089
## 3: 0.089
## 4: 0.089
## 5: 0.089
## ---
## 33373: 0.381
## 33374: 0.361
## 33375: 0.373
## 33376: 0.344
## 33377: 0.331ggplot2 makes it easy to visualise our data.
library(ggplot2)
library(dplyr)
rights |>
filter(entity %in% c("United Kingdom", "France", "United States")) |>
ggplot(aes(year, civ_libs_vdem_owid, colour = entity)) +
geom_line()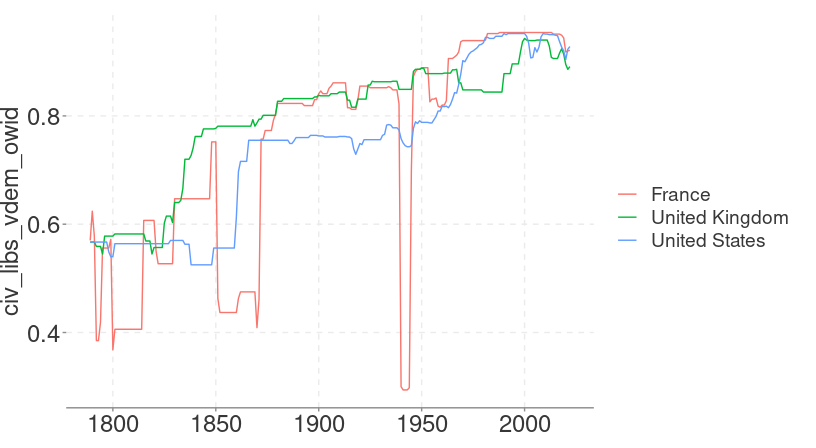
You can quickly download world covid-19 data, including vaccination
rates, using owid_covid().
covid <- owid_covid()
str(covid)
## Classes 'owid', 'data.table' and 'data.frame': 351765 obs. of 67 variables:
## $ iso_code : chr "AFG" "AFG" "AFG" "AFG" ...
## $ continent : chr "Asia" "Asia" "Asia" "Asia" ...
## $ location : chr "Afghanistan" "Afghanistan" "Afghanistan" "Afghanistan" ...
## $ date : IDate, format: "2020-01-03" "2020-01-04" ...
## $ total_cases : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_cases : num 0 0 0 0 0 0 0 0 0 0 ...
## $ new_cases_smoothed : num NA NA NA NA NA 0 0 0 0 0 ...
## $ total_deaths : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_deaths : num 0 0 0 0 0 0 0 0 0 0 ...
## $ new_deaths_smoothed : num NA NA NA NA NA 0 0 0 0 0 ...
## $ total_cases_per_million : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_cases_per_million : num 0 0 0 0 0 0 0 0 0 0 ...
## $ new_cases_smoothed_per_million : num NA NA NA NA NA 0 0 0 0 0 ...
## $ total_deaths_per_million : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_deaths_per_million : num 0 0 0 0 0 0 0 0 0 0 ...
## $ new_deaths_smoothed_per_million : num NA NA NA NA NA 0 0 0 0 0 ...
## $ reproduction_rate : num NA NA NA NA NA NA NA NA NA NA ...
## $ icu_patients : num NA NA NA NA NA NA NA NA NA NA ...
## $ icu_patients_per_million : num NA NA NA NA NA NA NA NA NA NA ...
## $ hosp_patients : num NA NA NA NA NA NA NA NA NA NA ...
## $ hosp_patients_per_million : num NA NA NA NA NA NA NA NA NA NA ...
## $ weekly_icu_admissions : num NA NA NA NA NA NA NA NA NA NA ...
## $ weekly_icu_admissions_per_million : num NA NA NA NA NA NA NA NA NA NA ...
## $ weekly_hosp_admissions : num NA NA NA NA NA NA NA NA NA NA ...
## $ weekly_hosp_admissions_per_million : num NA NA NA NA NA NA NA NA NA NA ...
## $ total_tests : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_tests : num NA NA NA NA NA NA NA NA NA NA ...
## $ total_tests_per_thousand : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_tests_per_thousand : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_tests_smoothed : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_tests_smoothed_per_thousand : num NA NA NA NA NA NA NA NA NA NA ...
## $ positive_rate : num NA NA NA NA NA NA NA NA NA NA ...
## $ tests_per_case : num NA NA NA NA NA NA NA NA NA NA ...
## $ tests_units : chr "" "" "" "" ...
## $ total_vaccinations : num NA NA NA NA NA NA NA NA NA NA ...
## $ people_vaccinated : num NA NA NA NA NA NA NA NA NA NA ...
## $ people_fully_vaccinated : num NA NA NA NA NA NA NA NA NA NA ...
## $ total_boosters : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_vaccinations : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_vaccinations_smoothed : num NA NA NA NA NA NA NA NA NA NA ...
## $ total_vaccinations_per_hundred : num NA NA NA NA NA NA NA NA NA NA ...
## $ people_vaccinated_per_hundred : num NA NA NA NA NA NA NA NA NA NA ...
## $ people_fully_vaccinated_per_hundred : num NA NA NA NA NA NA NA NA NA NA ...
## $ total_boosters_per_hundred : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_vaccinations_smoothed_per_million : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_people_vaccinated_smoothed : num NA NA NA NA NA NA NA NA NA NA ...
## $ new_people_vaccinated_smoothed_per_hundred: num NA NA NA NA NA NA NA NA NA NA ...
## $ stringency_index : num 0 0 0 0 0 0 0 0 0 0 ...
## $ population_density : num 54.4 54.4 54.4 54.4 54.4 ...
## $ median_age : num 18.6 18.6 18.6 18.6 18.6 18.6 18.6 18.6 18.6 18.6 ...
## $ aged_65_older : num 2.58 2.58 2.58 2.58 2.58 ...
## $ aged_70_older : num 1.34 1.34 1.34 1.34 1.34 ...
## $ gdp_per_capita : num 1804 1804 1804 1804 1804 ...
## $ extreme_poverty : num NA NA NA NA NA NA NA NA NA NA ...
## $ cardiovasc_death_rate : num 597 597 597 597 597 ...
## $ diabetes_prevalence : num 9.59 9.59 9.59 9.59 9.59 9.59 9.59 9.59 9.59 9.59 ...
## $ female_smokers : num NA NA NA NA NA NA NA NA NA NA ...
## $ male_smokers : num NA NA NA NA NA NA NA NA NA NA ...
## $ handwashing_facilities : num 37.7 37.7 37.7 37.7 37.7 ...
## $ hospital_beds_per_thousand : num 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 ...
## $ life_expectancy : num 64.8 64.8 64.8 64.8 64.8 ...
## $ human_development_index : num 0.511 0.511 0.511 0.511 0.511 0.511 0.511 0.511 0.511 0.511 ...
## $ population : num 41128772 41128772 41128772 41128772 41128772 ...
## $ excess_mortality_cumulative_absolute : num NA NA NA NA NA NA NA NA NA NA ...
## $ excess_mortality_cumulative : num NA NA NA NA NA NA NA NA NA NA ...
## $ excess_mortality : num NA NA NA NA NA NA NA NA NA NA ...
## $ excess_mortality_cumulative_per_million : num NA NA NA NA NA NA NA NA NA NA ...
## - attr(*, ".internal.selfref")=<externalptr>These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.