
The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.

Title: Higher-Level Interface of ‘torch’ Package to Auto-Train Neural Networks
Whether you’re generating neural network architectures expressions or
fitting/training actual models, {kindling} minimizes
boilerplate code while preserving {torch}. Since this
package uses {torch} as its backend, GPU/TPU devices also
supported.
{kindling} also bridges the gap between
{torch} and {tidymodels}. It works seamlessly
with {parsnip}, {recipes}, and
{workflows} to bring deep learning into your existing
{tidymodels} modeling pipeline. This enables a streamlined
interface for building, training, and tuning deep learning models within
the familiar {tidymodels} ecosystem.
{torch} expression{tidymodels}, {mlr3} for later) workflows and
pipelines{torch} tensors
You can install {kindling} on CRAN:
install.packages('kindling')Or install the development version from GitHub:
# install.packages("pak")
pak::pak("joshuamarie/kindling")
## devtools::install_github("joshuamarie/kindling") {kindling} is powered by R’s metaprogramming
capabilities through code generation. Generated
torch::nn_module() expressions power the training
functions, which in turn serve as engines for {tidymodels}
integration. This architecture gives you flexibility to work at whatever
abstraction level suits your task.
library(kindling)
#>
#> Attaching package: 'kindling'
#> The following object is masked from 'package:base':
#>
#> argsBefore starting, you need to install LibTorch, the backend of PyTorch
which also the backend of {torch} R package:
torch::install_torch()torch::nn_moduleAt the lowest level, you can generate raw
torch::nn_module code for maximum customization. Functions
ending with _generator return unevaluated expressions you
can inspect, modify, or execute.
Here’s how to generate a feedforward network specification:
ffnn_generator(
nn_name = "MyFFNN",
hd_neurons = c(64, 32, 16),
no_x = 10,
no_y = 1,
activations = 'relu'
)
#> torch::nn_module("MyFFNN", initialize = function ()
#> {
#> self$fc1 = torch::nn_linear(10, 64, bias = TRUE)
#> self$fc2 = torch::nn_linear(64, 32, bias = TRUE)
#> self$fc3 = torch::nn_linear(32, 16, bias = TRUE)
#> self$out = torch::nn_linear(16, 1, bias = TRUE)
#> }, forward = function (x)
#> {
#> x = self$fc1(x)
#> x = torch::nnf_relu(x)
#> x = self$fc2(x)
#> x = torch::nnf_relu(x)
#> x = self$fc3(x)
#> x = torch::nnf_relu(x)
#> x = self$out(x)
#> x
#> })This creates a three-hidden-layer network (64 - 32 - 16 neurons) that takes 10 inputs and produces 1 output. Each hidden layer uses ReLU activation, while the output layer remains “untransformed”.
Skip the code generation and train models directly with your data.
This approach handles all the {torch} boilerplate when
training the models internally.
Let’s classify iris species:
model = ffnn(
Species ~ .,
data = iris,
hidden_neurons = c(10, 15, 7),
activations = act_funs(relu, softshrink = args(lambd = 0.5), elu),
loss = "cross_entropy",
epochs = 100
)
model======================= Feedforward Neural Networks (MLP) ======================
-- FFNN Model Summary ----------------------------------------------------------
-----------------------------------------------------------------------
NN Model Type : FFNN n_predictors : 4
Number of Epochs : 100 n_response : 3
Hidden Layer Units : 10, 15, 7 reg. : None
Number of Hidden Layers : 3 Device : cpu
Pred. Type : classification :
-----------------------------------------------------------------------
-- Activation function ---------------------------------------------------------
-------------------------------------------------
1st Layer {10} : relu
2nd Layer {15} : softshrink(lambd = 0.5)
3rd Layer {7} : elu
Output Activation : No act function applied
-------------------------------------------------Evaluate the prediction through predict(). The
predict() method is extended for fitted models through its
newdata argument.
Two kinds of predict() usage:
Without newdata predictions is the
default to the parent data frame.
predict(model) |>
(\(x) table(actual = iris$Species, predicted = x))()
#> predicted
#> actual setosa versicolor virginica
#> setosa 50 0 0
#> versicolor 0 46 4
#> virginica 0 2 48With newdata simply pass the new
data frame as the new reference.
sample_iris = dplyr::slice_sample(iris, n = 10, by = Species)
predict(model, newdata = sample_iris) |>
(\(x) table(actual = sample_iris$Species, predicted = x))()
#> predicted
#> actual setosa versicolor virginica
#> setosa 10 0 0
#> versicolor 0 10 0
#> virginica 0 1 9Work with neural networks just like any other {parsnip}
model. This unlocks the entire {tidymodels} toolkit for
preprocessing, cross-validation, and model evaluation.
# library(kindling)
# library(parsnip)
# library(yardstick)
box::use(
kindling[mlp_kindling, rnn_kindling, act_funs, args],
parsnip[fit, augment],
yardstick[metrics],
mlbench[Ionosphere] # data(Ionosphere, package = "mlbench")
)
ionosphere_data = Ionosphere[, -2]
# Train a feedforward network with parsnip
mlp_kindling(
mode = "classification",
hidden_neurons = c(128, 64),
activations = act_funs(relu, softshrink = args(lambd = 0.5)),
epochs = 100
) |>
fit(Class ~ ., data = ionosphere_data) |>
augment(new_data = ionosphere_data) |>
metrics(truth = Class, estimate = .pred_class)
#> # A tibble: 2 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 accuracy binary 0.989
#> 2 kap binary 0.975
# Or try a recurrent architecture (demonstrative example with tabular data)
rnn_kindling(
mode = "classification",
hidden_neurons = c(128, 64),
activations = act_funs(relu, elu),
epochs = 100,
rnn_type = "gru"
) |>
fit(Class ~ ., data = ionosphere_data) |>
augment(new_data = ionosphere_data) |>
metrics(truth = Class, estimate = .pred_class)
#> # A tibble: 2 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 accuracy binary 0.641
#> 2 kap binary 0The package has integration with {tidymodels}, so it
supports hyperparameter tuning via {tune} with searchable
parameters.
The current searchable parameters under {kindling}:
The searchable parameters outside from {kindling},
i.e. under {dials} package such as
learn_rate() also supported.
Here’s an example:
# library(tidymodels)
box::use(
kindling[
mlp_kindling, hidden_neurons, activations, output_activation, grid_depth
],
parsnip[fit, augment],
recipes[recipe],
workflows[workflow, add_recipe, add_model],
rsample[vfold_cv],
tune[tune_grid, tune, select_best, finalize_workflow],
dials[grid_random],
yardstick[accuracy, roc_auc, metric_set, metrics]
)
mlp_tune_spec = mlp_kindling(
mode = "classification",
hidden_neurons = tune(),
activations = tune(),
output_activation = tune()
)
iris_folds = vfold_cv(iris, v = 3)
nn_wf = workflow() |>
add_recipe(recipe(Species ~ ., data = iris)) |>
add_model(mlp_tune_spec)
nn_grid_depth = grid_depth(
hidden_neurons(c(32L, 128L)),
activations(c("relu", "elu")),
output_activation(c("sigmoid", "linear")),
n_hlayer = 2,
size = 10,
type = "latin_hypercube"
)
# This is supported but limited to 1 hidden layer only
## nn_grid = grid_random(
## hidden_neurons(c(32L, 128L)),
## activations(c("relu", "elu")),
## output_activation(c("sigmoid", "linear")),
## size = 10
## )
nn_tunes = tune::tune_grid(
nn_wf,
iris_folds,
grid = nn_grid_depth
# metrics = metric_set(accuracy, roc_auc)
)
best_nn = select_best(nn_tunes)
final_nn = finalize_workflow(nn_wf, best_nn)
# Last run: 4 - 91 (relu) - 3 (sigmoid) units
final_nn_model = fit(final_nn, data = iris)
final_nn_model |>
augment(new_data = iris) |>
metrics(truth = Species, estimate = .pred_class)
#> # A tibble: 2 × 3
#> .metric .estimator .estimate
#> <chr> <chr> <dbl>
#> 1 accuracy multiclass 0.667
#> 2 kap multiclass 0.5Resampling strategies from {rsample} will enable robust
cross-validation workflows, orchestrated through the {tune}
and {dials} APIs.
{kindling} integrates with established variable
importance methods from {NeuralNetTools} and
{vip} to interpret trained neural networks. Two primary
algorithms are available:
Garson’s Algorithm
garson(model, bar_plot = FALSE)
#> x_names y_names rel_imp
#> 1 Sepal.Width y 29.04598
#> 2 Petal.Width y 27.50590
#> 3 Sepal.Length y 24.20982
#> 4 Petal.Length y 19.23830Olden’s Algorithm
olden(model, bar_plot = FALSE)
#> x_names y_names rel_imp
#> 1 Sepal.Width y 0.56231712
#> 2 Petal.Width y -0.51113650
#> 3 Petal.Length y -0.29761552
#> 4 Sepal.Length y -0.06857191For users working within the {tidymodels} ecosystem,
{kindling} models work seamlessly with the
{vip} package:
box::use(
vip[vi, vip]
)
vi(model) |>
vip()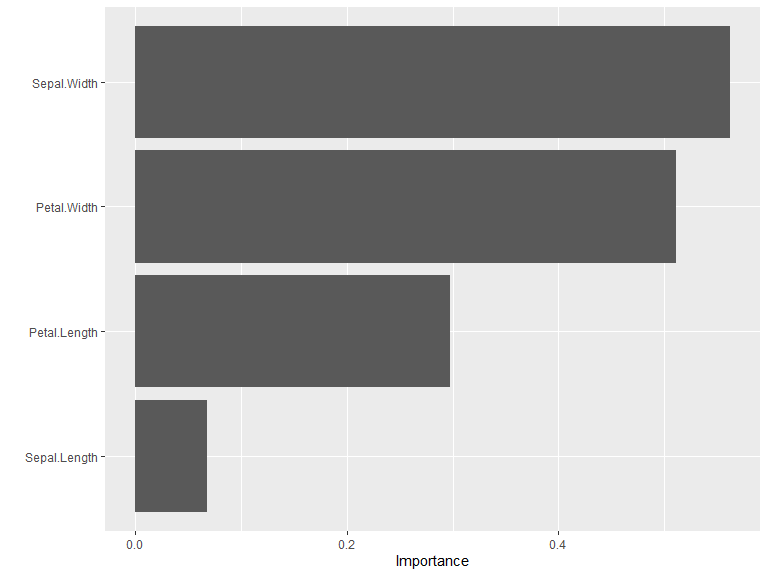
Note: Weight caching increases memory usage proportional to network size. Only enable it when you plan to compute variable importance multiple times on the same model.
Falbel D, Luraschi J (2023). torch: Tensors and Neural Networks with ‘GPU’ Acceleration. R package version 0.13.0, https://torch.mlverse.org, https://github.com/mlverse/torch.
Wickham H (2019). Advanced R, 2nd edition. Chapman and Hall/CRC. ISBN 978-0815384571, https://adv-r.hadley.nz/.
Goodfellow I, Bengio Y, Courville A (2016). Deep Learning. MIT Press. https://www.deeplearningbook.org/.
MIT + file LICENSE
Please note that the kindling project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.