The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
{guescini} is an R data package that provides real-time
PCR raw fluorescence data by Guescini et al. (2008) in tidy format.
Install {guescini} from CRAN:
# Install from CRAN
install.packages("guescini")Guescini et al. (2008) explored the effect of amplification inhibition on qPCR quantification. Two systems were devised to alter the amplification efficiency:
The raw fluorescence data associated with the decreasing of the
amplification mix is provided as the data set amp_mix_perc;
the data obtained with increasing concentrations of IgG is provided as
IgG_inhibition.
The data set amp_mix_perc corresponds to a set of
amplification runs where the MT-ND1 gene is amplified in reactions
having the same initial amount of DNA but different amounts of SYBR
Green I Master mix. A standard curve was performed over a wide range of
input DNA (\(3.14 \times 10^7\ \text{thru}\
3.14 \times 10^1\)) in the presence of optimal amplification
conditions (100% amplification mix), while the unknowns were run in the
presence of the same starting DNA amounts but with amplification mix
quantities ranging from 60% to 100%.
library(guescini)
amp_mix_perc
#> # A tibble: 21,000 × 12
#> plate well dye target sample_type run replicate amp_mix_perc copies
#> <fct> <fct> <fct> <fct> <fct> <fct> <fct> <dbl> <int>
#> 1 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 2 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 3 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 4 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 5 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 6 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 7 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 8 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 9 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> 10 <NA> <NA> SYBR MT-ND1 std 1 1 1 31400000
#> # ℹ 20,990 more rows
#> # ℹ 3 more variables: dilution <int>, cycle <int>, fluor <dbl>
amp_mix_perc %>%
ggplot(mapping = aes(
x = cycle,
y = fluor,
group = interaction(run, amp_mix_perc, copies),
col = format(copies, big.mark = ",", scientific = FALSE)
)) +
geom_line(linewidth = 0.2) +
geom_point(size = 0.2) +
labs(y = "Raw fluorescence", colour = "No. of copies", title = "Seven-point 10-fold dilution series amplification mix percentage") +
guides(color = guide_legend(override.aes = list(linewidth = 0.5), reverse = TRUE)) +
facet_wrap(vars(amp_mix_perc))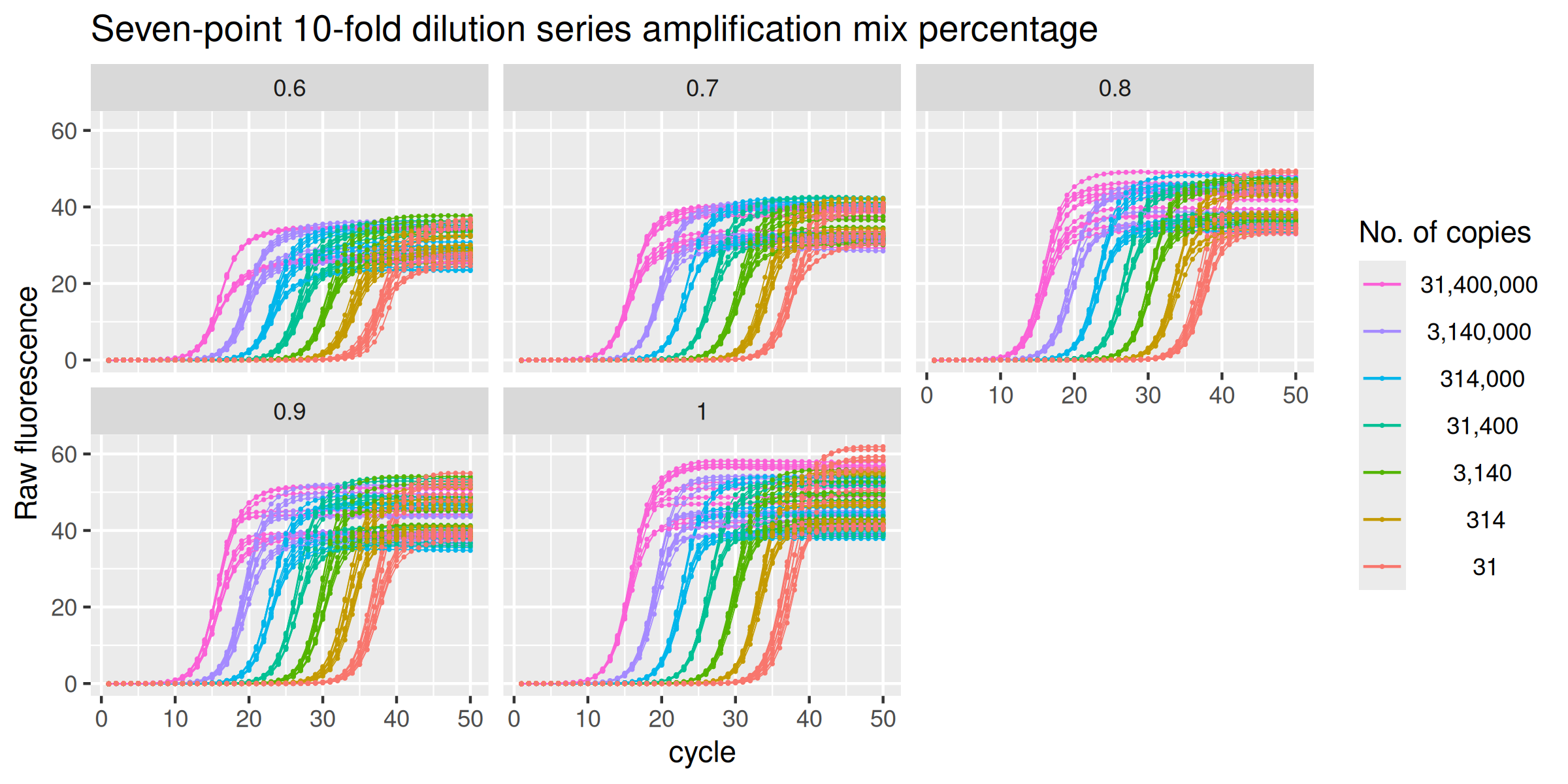
The data set IgG_inhibition provides those runs
performed in the presence of an optimal amplification reaction mix added
with serial dilutions of IgG (0.0 - 2 ug/ml) thus acting as the
inhibitory agent.
IgG_inhibition %>%
ggplot(mapping = aes(
x = cycle,
y = fluor,
group = interaction(IgG_conc, replicate),
col = paste(as.character(IgG_conc), "ug/ml")
)) +
geom_line(linewidth = 0.5) +
geom_point(size = 0.5) +
labs(y = "Raw fluorescence", colour = "IgG concentration", title = "Serial dilutions of IgG (PCR inhibitor)") +
guides(color = guide_legend(override.aes = list(linewidth = 0.5)))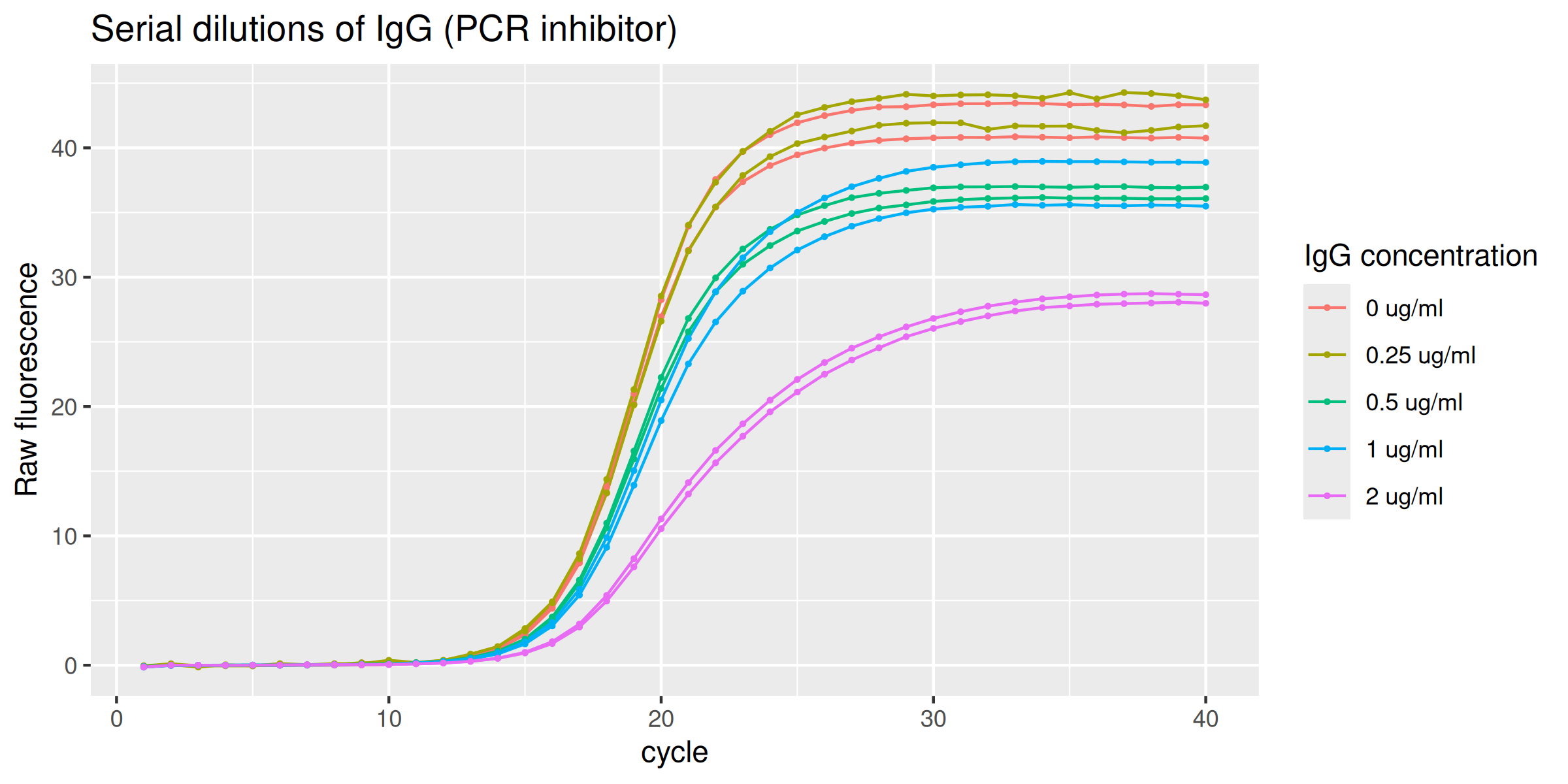
Please note that the guescini project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
Michele Guescini, Davide Sisti, Marco BL Rocchi, Laura Stocchi and Vilberto Stocchi. A new real-time PCR method to overcome significant quantitative inaccuracy due to slight amplification inhibition. BMC Bioinformatics 9:326 (2008). doi: 10.1186/1471-2105-9-326.
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.