The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
FDR functions for permutation-based estimators, including pi0 as well as FDR confidence intervals. The confidence intervals account for dependencies between tests by the incorporation of an overdispersion parameter, which is estimated from the permuted data. — From the package description
You can install fdrci from GitHub with:
# install.packages("devtools")
devtools::install_github("USCbiostats/fdrci")This is a basic example which shows you how to solve a common problem:
library(fdrci)
ss = 100
nvar = 100
X = as.data.frame(matrix(rnorm(ss*nvar),nrow=ss,ncol=nvar))
Y = as.data.frame(matrix(rnorm(ss*nvar),nrow=ss,ncol=nvar))
nperm = 10
myanalysis = function(X,Y){
ntests = ncol(X)
rslts = as.data.frame(matrix(NA,nrow=ntests,ncol=2))
names(rslts) = c("ID","pvalue")
rslts[,"ID"] = 1:ntests
for(i in 1:ntests){
fit = cor.test(X[,i],Y[,i],na.action="na.exclude",
alternative="two.sided",method="pearson")
rslts[i,"pvalue"] = fit$p.value
}
return(rslts)
} # End myanalysis
# Generate observed results
obs = myanalysis(X,Y)
# Generate permuted results
perml = vector('list',nperm)
for(p_ in 1:nperm){
X1 = X[order(runif(nvar)),]
perml[[p_]] = myanalysis(X1,Y)
}
# FDR results table
myfdrtbl = fdrTbl(obs$pvalue,perml,"pvalue",nvar,0,3)
# Plot results
FDRplot(myfdrtbl,0,3,annot="A. An Example")
#> Warning: Removed 13 rows containing missing values (geom_path).
#> Warning: Removed 13 rows containing missing values (geom_point).
#> Warning: Removed 13 rows containing missing values (geom_text).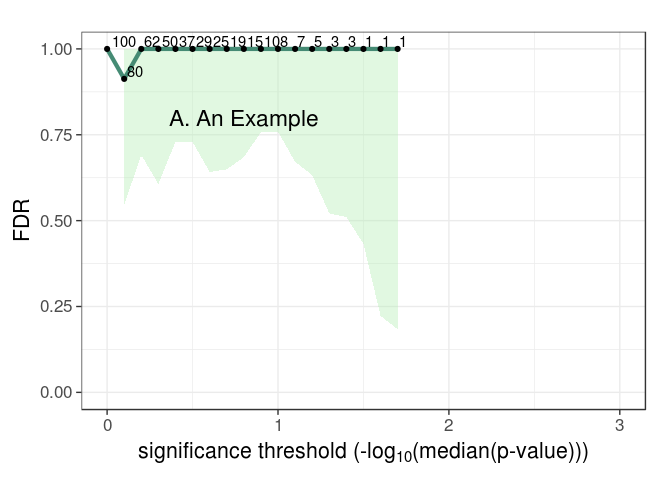
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.