The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The First-order Integer-valued Autoregressive (INAR(1)) model with zero-inflated (ZI-INAR(1)) and hurdle (H-INAR(1)) innovations is widely used in studying integer-valued time-series data, such as crime count and heatwave frequency. This work implemented the INAR(1) models in Stan.
You can install ZIHINAR1 from GitHub with:
remotes::install_github("fushengyy/ZIHINAR1")\[Available Soon\] Or you can install the released version of HeckmanStan from CRAN with:
install.packages("ZIHINAR1")The package contains main function named get_stanfit().
stan_fit <- get_stanfit(mod_type, distri, y, n_pred = 4,
thin = 2, chains = 1, iter = 2000, warmup = iter/2,
seed = NA)mod_type: Character string indicating the model type. Use “zi” for zero-inflated models and “h” for hurdle models.
distri: Character string specifying the distribution. Options are “poi” for Poisson or “nb” for Negative Binomial.
y: A numeric vector of integers representing the observed data.
n_pred: Integer specifying the number of time points for future predictions (default is 4).
thin: Integer indicating the thinning interval for Stan sampling (default is 2).
chains: Integer specifying the number of Markov chains to run (default is 1).
iter: Integer specifying the total number of iterations per chain (default is 2000).
warmup: Integer specifying the number of warmup iterations per chain (default is iter/2).
seed: Numeric seed for reproducibility (default is NA).
The following are examples showing how to fit the INAR(1) model when data is generated from a zero-inflated Negative Binomial (ZINB) distribution.
library(ZIHINAR1)
y_data <- data_simu(n = 100, alpha = 0.5, rho = 0.3, theta = c(5, 2),
mod_type = "zi", distri = "nb")
stan_fit <- get_stanfit(mod_type = "zi", distri = "nb", y = y_data, n_pred = 5,
iter = 2000, chains = 1, warmup = 500,
thin = 2, seed = 42)
#>
#> SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 0.002477 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 24.77 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 1: Iteration: 501 / 2000 [ 25%] (Sampling)
#> Chain 1: Iteration: 700 / 2000 [ 35%] (Sampling)
#> Chain 1: Iteration: 900 / 2000 [ 45%] (Sampling)
#> Chain 1: Iteration: 1100 / 2000 [ 55%] (Sampling)
#> Chain 1: Iteration: 1300 / 2000 [ 65%] (Sampling)
#> Chain 1: Iteration: 1500 / 2000 [ 75%] (Sampling)
#> Chain 1: Iteration: 1700 / 2000 [ 85%] (Sampling)
#> Chain 1: Iteration: 1900 / 2000 [ 95%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 3.767 seconds (Warm-up)
#> Chain 1: 9.449 seconds (Sampling)
#> Chain 1: 13.216 seconds (Total)
#> Chain 1:
get_est(distri = "nb", stan_fit = stan_fit)
#>
#>
#> Table: Parameter Estimates
#>
#> Mean SD Median Q2.5 Q97.5 Rhat 95%_HPD_Lower 95%_HPD_Upper
#> ------- ------- ------- ------- ------- ------- ------- -------------- --------------
#> alpha 0.5434 0.0407 0.5450 0.4620 0.6123 1.0010 0.4605 0.6113
#> rho 0.2573 0.1131 0.2506 0.0404 0.4742 1.0006 0.0437 0.4753
#> lambda 4.9955 0.7772 4.9750 3.6025 6.5105 0.9987 3.6044 6.5105
#> phi 2.2278 1.2160 1.9962 0.7745 4.9245 0.9993 0.5618 4.3624
get_mod_sel(y = y_data, mod_type = "zi", distri = "nb", stan_fit = stan_fit)
#>
#>
#> Table: Model Selection Criteria
#>
#> EAIC EBIC DIC WAIC1 WAIC2
#> --------- --------- --------- --------- ---------
#> 554.6341 565.0146 555.8627 550.0862 550.3367
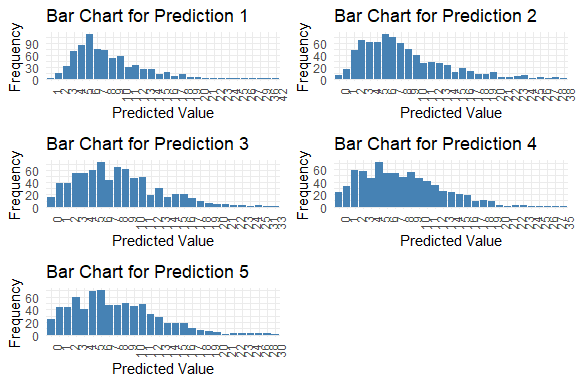
get_pred(stan_fit = stan_fit)
#>
#>
#> Table: Summary of Predictions
#>
#> Mode Median IQR Min Max
#> --------- ----- ------- ---- ---- ----
#> y_pred.1 6 7.0 5 1 42
#> y_pred.2 6 7.0 7 0 38
#> y_pred.3 6 7.5 7 0 33
#> y_pred.4 5 7.0 7 0 35
#> y_pred.5 6 7.0 7 0 30
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.