The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The package provides an R interface for processing concentration-response datasets using Curvep, a response noise filtering algorithm. The algorithm was described in the publications (Sedykh A et al. (2011) doi:10.1289/ehp.1002476 and Sedykh A (2016) doi:10.1007/978-1-4939-6346-1_14).
Other parametric fitting approaches (e.g., Hill equation) are also adopted for ease of comparison. 3-parameter Hill equation from original tcpl package (Filer DL et al., doi:10.1093/bioinformatics/btw680) and 4-parameter Hill equation from Curve Class2 approach (Wang Y et al., doi:10.2174/1875397301004010057) are available.
Also, methods for calculating the confidence interval around the activity metrics are also provided. The methods are based on the bootstrap approach to simulate the datasets (Hsieh J-H et al. doi:10.1093/toxsci/kfy258). The simulated datasets can be used to derive the baseline noise threshold in an assay endpoint. This threshold is critical in the toxicological studies to derive the point-of-departure (POD).
# the development version from GitHub:
# install.packages("devtools")
devtools::install_github("moggces/Rcurvep")
devtools::install_github("moggces/Rcurvep", dependencies = TRUE, build_vignettes = TRUE)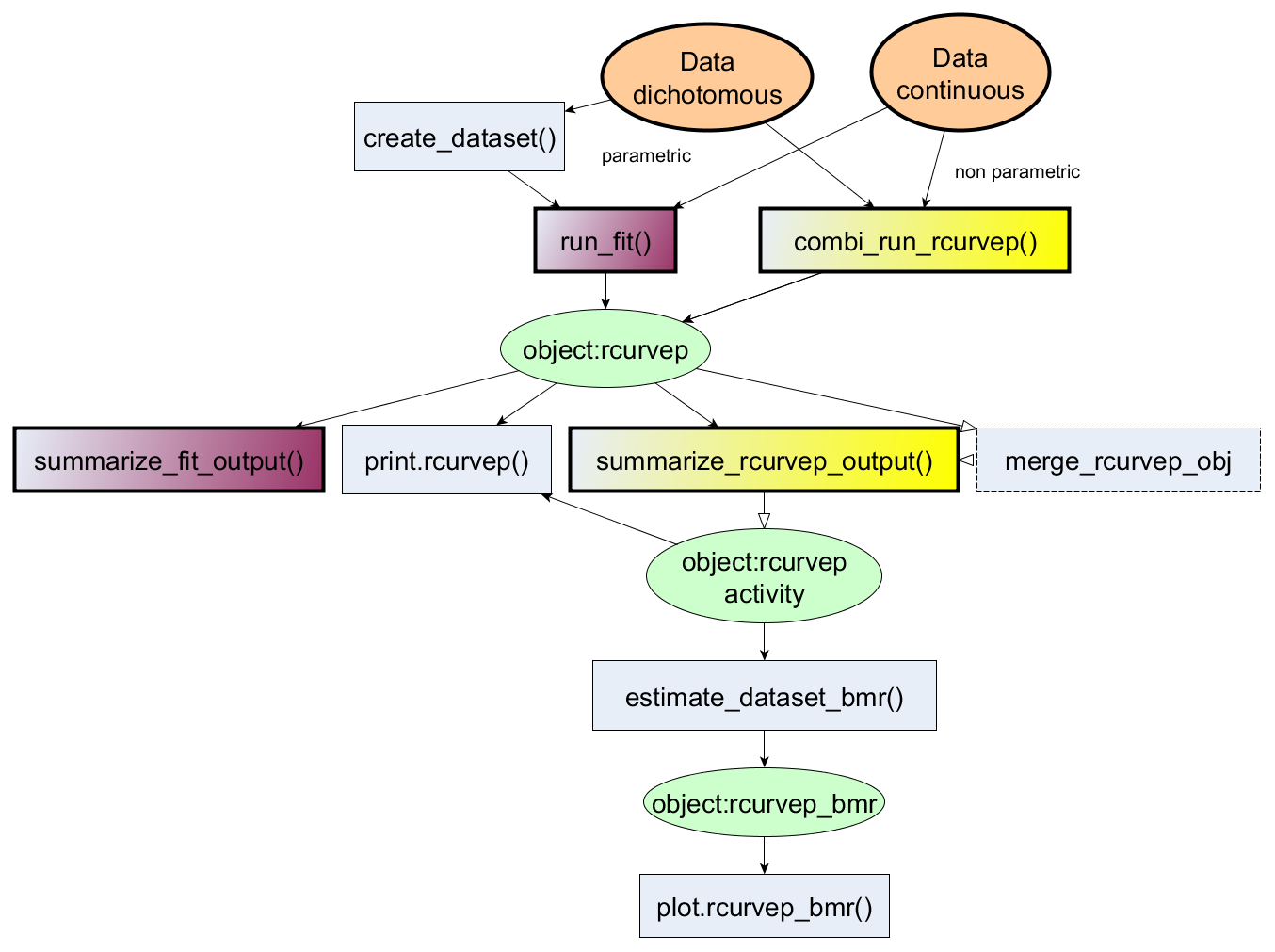
library(Rcurvep)
data("zfishbeh")
out_curvep <- combi_run_rcurvep(zfishbeh, TRSH = 30) # using Curvep with BMR = 30
out_fit1 <- run_fit(zfishbeh, modls = "cc2") # using Curve Class2 4-parameter hill
out_fit2 <- run_fit(zfishbeh, modls = c("cnst", "hill")) # using tcpl 3-parameter hill + constant modeldata("zfishdev_act")
out_bmr <- estimate_dataset_bmr(zfishdev_act)## $`1`
To learn more about Rcurvep, start with the vignettes:
browseVignettes(package = "Rcurvep")
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.