The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
The REDI package implements the Robust Exponential Decreasing Index (REDI). It represents a measure of cumulated workload, is robust to missing data and provides control of the decreasing influence of the workload over time.
REDI provides various functions to format data, compute REDI and visualise results in a simple and convenient way.
Issa Moussa, Arthur Leroy et al. (2019): Robust Exponential Decreasing Index (REDI): adaptive and robust method for computing cumulated workload. BMJ Open Sport & Exercise Medicine, https://bmjopensem.bmj.com/content/bmjosem/5/1/e000573.full.pdf.
You can install the development version of REDI from GitHub with:
#install.packages("devtools")
devtools::install_github("Grenouil/REDI")Here is a basic example on how to simulate a dataset with the adequate format, compute REDI values over time and display results.
library(REDI)
set.seed(42)
## Generate a synthetic dataset with the correct format
data = simu_db()
data
#> # A tibble: 366 x 2
#> Input Output
#> <date> <dbl>
#> 1 2022-01-01 54.3
#> 2 2022-01-02 NA
#> 3 2022-01-03 51.1
#> 4 2022-01-04 NA
#> 5 2022-01-05 NA
#> 6 2022-01-06 49.7
#> 7 2022-01-07 54.8
#> 8 2022-01-08 NA
#> 9 2022-01-09 NA
#> 10 2022-01-10 NA
#> # i 356 more rows
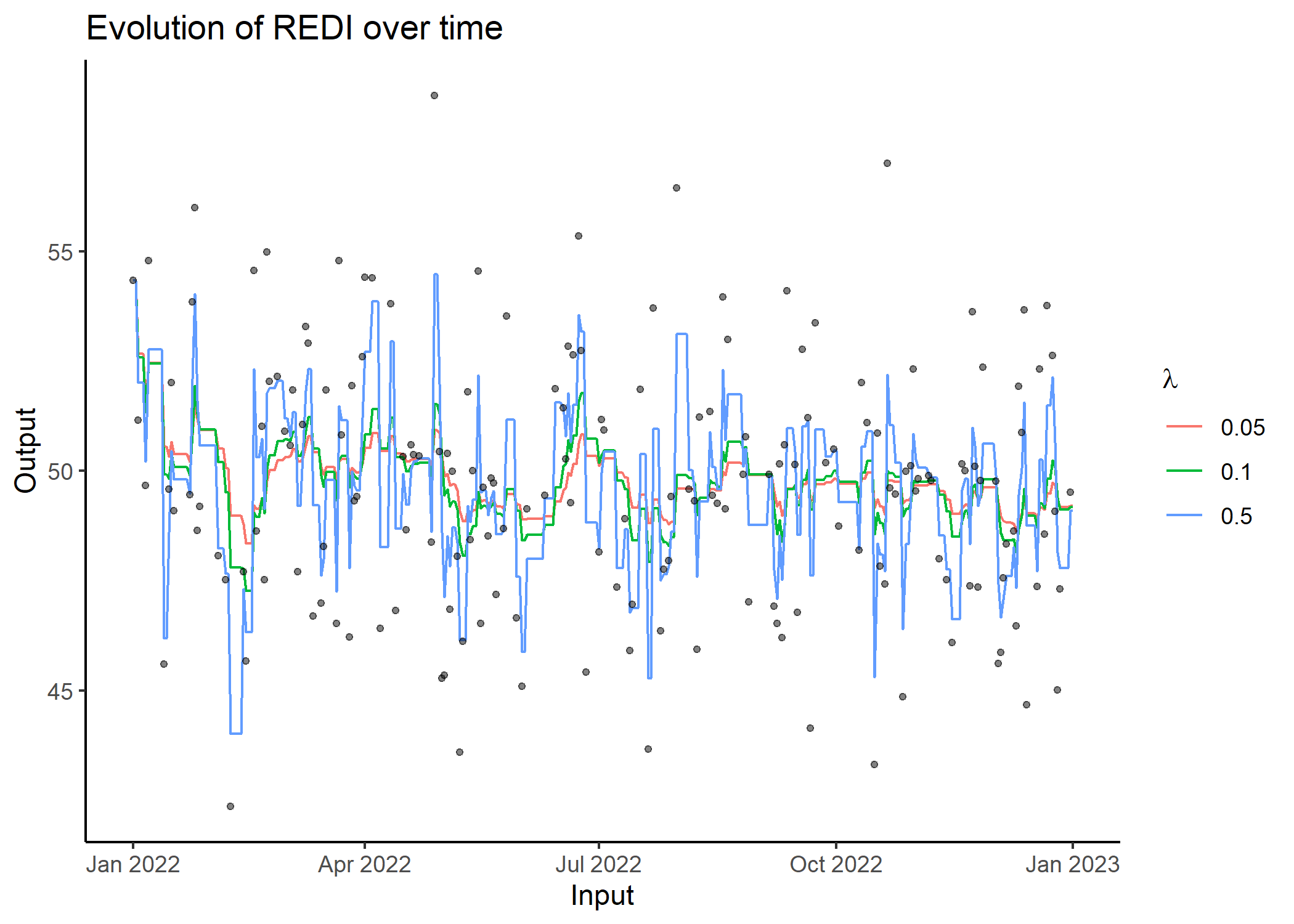
## Compute REDI over all observations for different Lambda values and display results
res = redi(data)
res
#> # A tibble: 1,098 x 4
#> Input Output REDI Lambda
#> <date> <dbl> <dbl> <dbl>
#> 1 2023-01-01 NA 49.2 0.05
#> 2 2022-12-31 49.5 49.2 0.05
#> 3 2022-12-30 NA 49.2 0.05
#> 4 2022-12-29 NA 49.2 0.05
#> 5 2022-12-28 NA 49.2 0.05
#> 6 2022-12-27 47.3 49.2 0.05
#> 7 2022-12-26 45.0 49.3 0.05
#> 8 2022-12-25 49.1 49.7 0.05
#> 9 2022-12-24 52.6 49.7 0.05
#> 10 2022-12-23 NA 49.5 0.05
#> # i 1,088 more rowsFor a advanced use of the package, here is a quick step-by-step guide.
In order to test different features of the package, the
simu_db() function is provided as a handy way to generate
synthetic data with the correct format for subsequent REDI
computations.
set.seed(42)
## Generate a synthetic dataset, containing dates (Inputs) from '2022-01-01' to '2023-01-01' and observations (Outputs) following a Gaussian distribution (mean = 50, var = 10), with 50% of missing values.
simu_data <- simu_db(start_date = '2022-01-01',
end_date = '2023-01-01',
by = 'day',
output_distrib = 'Gaussian',
ratio_missing = 0.5,
mean = 50,
var = 10)
simu_data
#> # A tibble: 366 x 2
#> Input Output
#> <date> <dbl>
#> 1 2022-01-01 54.3
#> 2 2022-01-02 NA
#> 3 2022-01-03 51.1
#> 4 2022-01-04 NA
#> 5 2022-01-05 NA
#> 6 2022-01-06 49.7
#> 7 2022-01-07 54.8
#> 8 2022-01-08 NA
#> 9 2022-01-09 NA
#> 10 2022-01-10 NA
#> # i 356 more rowsAs displayed above, any dataset processed in REDI should provide 2
columns: one corresponding to Input values (e.g.
time) and another to Output values (e.g.
workload).
format_data()However, a real-life dataset will generally not have the correct
format to compute REDI directy. Therefore, the
format_data() function is designed to help with this
process by identifying the columns corresponding to the Input (e.g the
date) and the Output (e.g. the workload) variables. The Input column
should be defined with a correct Date type. The function
will automatically identify missing values between each observations,
considering the by argument as the time increment (for
instance with ‘day’, the default, each day between two observed dates is
considered missing). Finally, Output values for duplicated Input values
can be summarised according to the summarise_duplicate
argument.
## Create a dummy real-life dataset
raw_db <- data.frame(
'Var1' = 1:100,
'Var2' = rnorm(n = 100, mean = 50, sd = 10),
'Var3' = c(
as.Date("2022/1/1"),
seq(from = as.Date("2022/1/3"), by = "day", length.out = 99)
)
)
head(raw_db)
#> Var1 Var2 Var3
#> 1 1 58.84844 2022-01-01
#> 2 2 51.63431 2022-01-03
#> 3 3 41.39630 2022-01-04
#> 4 4 59.74555 2022-01-05
#> 5 5 43.81340 2022-01-06
#> 6 6 44.72518 2022-01-07
## Convert the dataset to the correct format (adding the missing data point on 2022-01-02)
db <- format_data(
data = raw_db,
input = 'Var3',
output = 'Var2',
by = 'day',
format = '%Y%m%d'
)
db
#> # A tibble: 101 x 2
#> Input Output
#> <date> <dbl>
#> 1 2022-01-01 58.8
#> 2 2022-01-02 NA
#> 3 2022-01-03 51.6
#> 4 2022-01-04 41.4
#> 5 2022-01-05 59.7
#> 6 2022-01-06 43.8
#> 7 2022-01-07 44.7
#> 8 2022-01-08 32.4
#> 9 2022-01-09 45.0
#> 10 2022-01-10 46.8
#> # i 91 more rowscompute_redi()To compute a single REDI value, simply use the
compute_redi() function. It will correspond to the REDI
value for the most recent observed Input in the dataset, using all data
from the past. Feel free to adapt the \(\lambda\) coefficient, controlling the
exponential decay of weights over time, depending on the context.
## Compute REDI at the current date (2022-04-11 in this example)
compute_redi(db, coef = 0.05)
#> [1] 49.85942
max(db$Input)
#> [1] "2022-04-11"loop_redi()To sequentially compute REDI for all Input values in the
dataset with speed-up vectorised operations, one can use the
loop_redi() function.
## Apply loop_redi() to tcompute REDI for all dates in the dataset
db_redi <- loop_redi(data = db, coef = 0.5)
db_redi
#> # A tibble: 101 x 4
#> Input Output REDI Lambda
#> <date> <dbl> <dbl> <dbl>
#> 1 2022-04-11 50.1 51.4 0.5
#> 2 2022-04-10 63.9 52.3 0.5
#> 3 2022-04-09 40.9 44.9 0.5
#> 4 2022-04-08 48.3 47.4 0.5
#> 5 2022-04-07 48.7 46.8 0.5
#> 6 2022-04-06 37.9 45.6 0.5
#> 7 2022-04-05 54.4 50.6 0.5
#> 8 2022-04-04 36.9 48.2 0.5
#> 9 2022-04-03 55.0 55.5 0.5
#> 10 2022-04-02 63.0 55.8 0.5
#> # i 91 more rowsplot_redi()The plot_redi() function is proposed to display the
results, and provides several options to personalise the graphs.
## Display results as time series of REDI values
plot_redi(redi = db_redi,
x_axis = 'Date',
y_axis = 'Workload',
plot_data = TRUE)
#> Warning: Removed 1 rows containing missing values (`geom_point()`).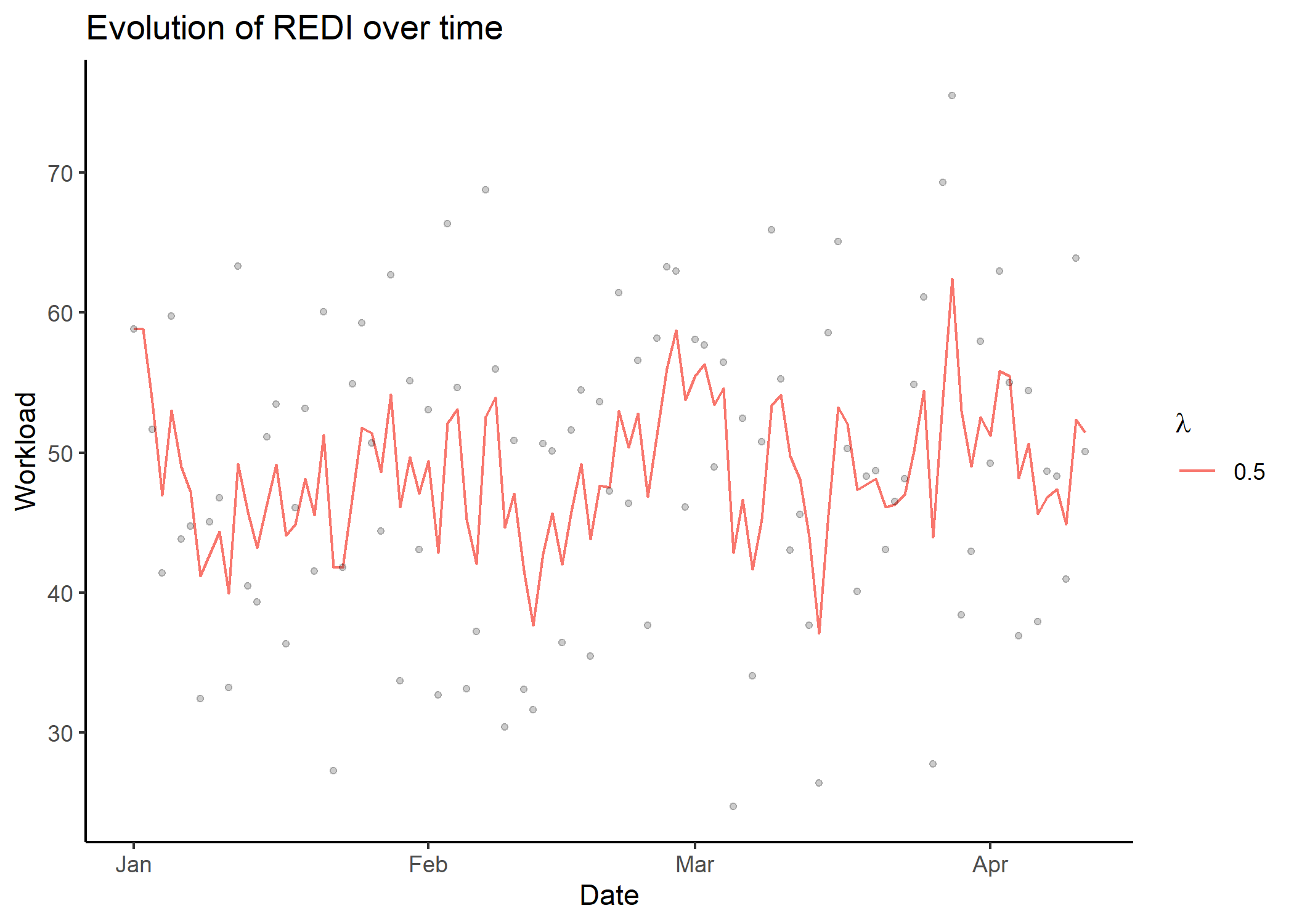
One can customise graphs by:
x_axis and y_axis;plot_data to FALSE.redi()As presented in the nutshell example, all the previous steps
(formatting, computations, plotting) are wrapped into the
redi() function. In addition to the arguments of the
previous functions, it is also possible to provide a vector to the
coef argument to display results for different \(\lambda\) values.
## Apply redi() on db and provide a vector of coefficients.
db_full_redi <- redi(data = db, coef = c(0.1, 0.2, 0.3), plot = TRUE)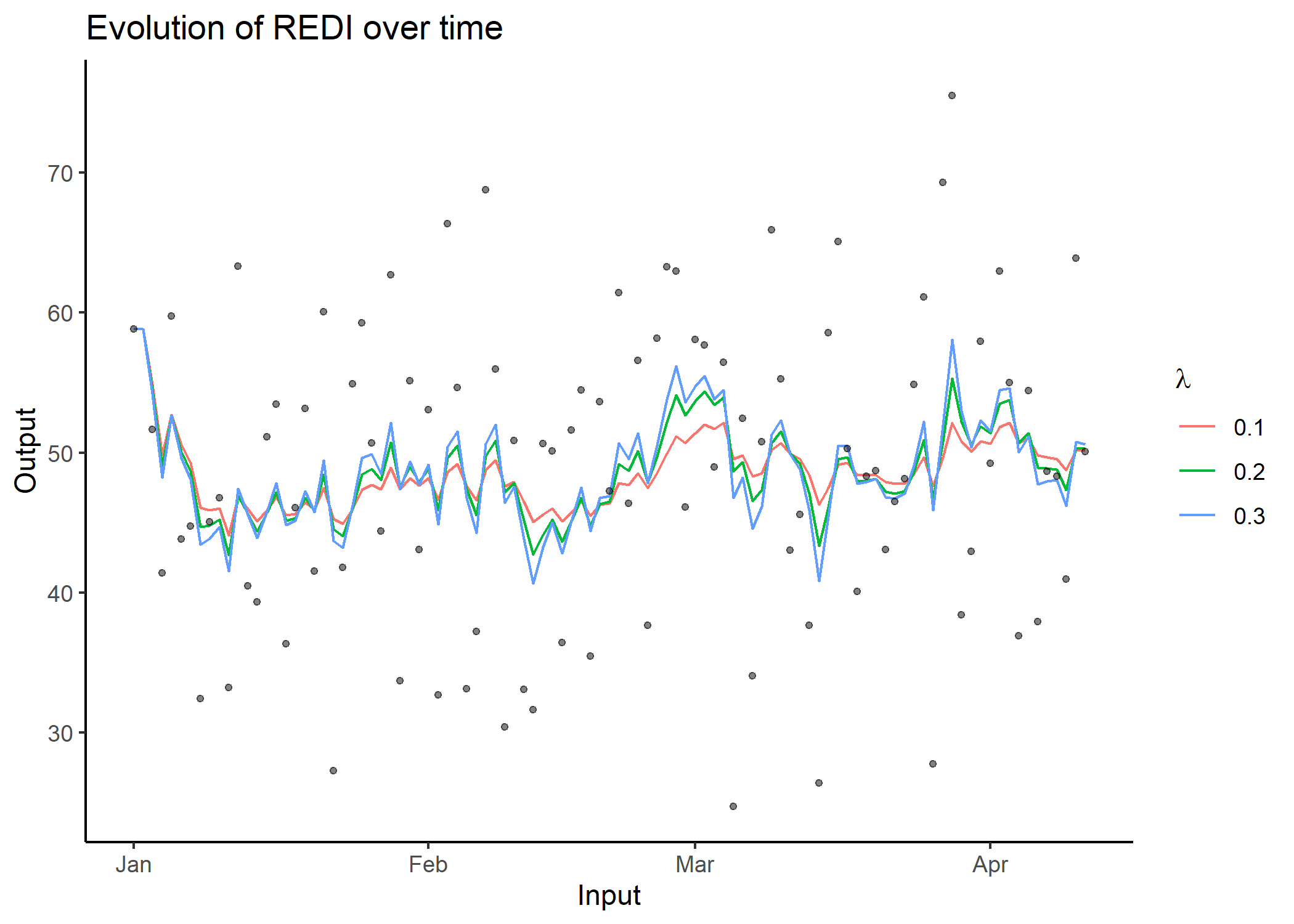
db_full_redi
#> # A tibble: 303 x 4
#> Input Output REDI Lambda
#> <date> <dbl> <dbl> <dbl>
#> 1 2022-04-11 50.1 50.2 0.1
#> 2 2022-04-10 63.9 50.2 0.1
#> 3 2022-04-09 40.9 48.7 0.1
#> 4 2022-04-08 48.3 49.5 0.1
#> 5 2022-04-07 48.7 49.7 0.1
#> 6 2022-04-06 37.9 49.8 0.1
#> 7 2022-04-05 54.4 51.0 0.1
#> 8 2022-04-04 36.9 50.7 0.1
#> 9 2022-04-03 55.0 52.1 0.1
#> 10 2022-04-02 63.0 51.8 0.1
#> # i 293 more rowsThese binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.