The hardware and bandwidth for this mirror is donated by dogado GmbH, the Webhosting and Full Service-Cloud Provider. Check out our Wordpress Tutorial.
If you wish to report a bug, or if you are interested in having us mirror your free-software or open-source project, please feel free to contact us at mirror[@]dogado.de.
PPtreeExt is an R package with extensons to the Projection Pursuit Tree (PPtree) algorithm to improve its performance in multi-class settings and under nonlinear separations. The PPtree classifier finds separations between classes based on linear combinations of variables by optimizing a projection pursuit index. One of its limitations is a rigid structure: the depth of a PPtree object is at most \(G\)-1, where \(G\) is the number of classes, with each class assigned to a single terminal node. The proposed modifications enhance predictive performance in multi-class contexts, particularly in situations involving outliers or asymmetries. The objective is to increase the classifier’s flexibility to handle more complex scenarios, while retaining interpretability. The package includes an interactive web application to explore the behavior of the original and modified PPtree classifiers under a variety of scenarios. This interactive tool played a key role in identifying limitations of the original algorithm and informing the design of the proposed modifications.
Install version from CRAN (not available yet!):
install.packages("PPTreeExt")Install the development version from GitHub:
# install.packages("devtools")
devtools::install_github("natydasilva/PPtreeExt")set.seed(234)
data(penguins)
penguins <- na.omit(penguins[, -c(2,7, 8)])
require(rsample)
penguins_spl <- rsample::initial_split(penguins, strata=species)
penguins_train <- training(penguins_spl)
penguins_test <- testing(penguins_spl)
penguins_ppt <- PPtreeExtclass(species~bill_len + bill_dep +
flipper_len + body_mass, data = penguins_train, PPmethod = "LDA", tot=nrow
(penguins_train), tol = 0.2 , srule = TRUE)
=============================================================
Projection Pursuit Classification Tree Extension result
=============================================================
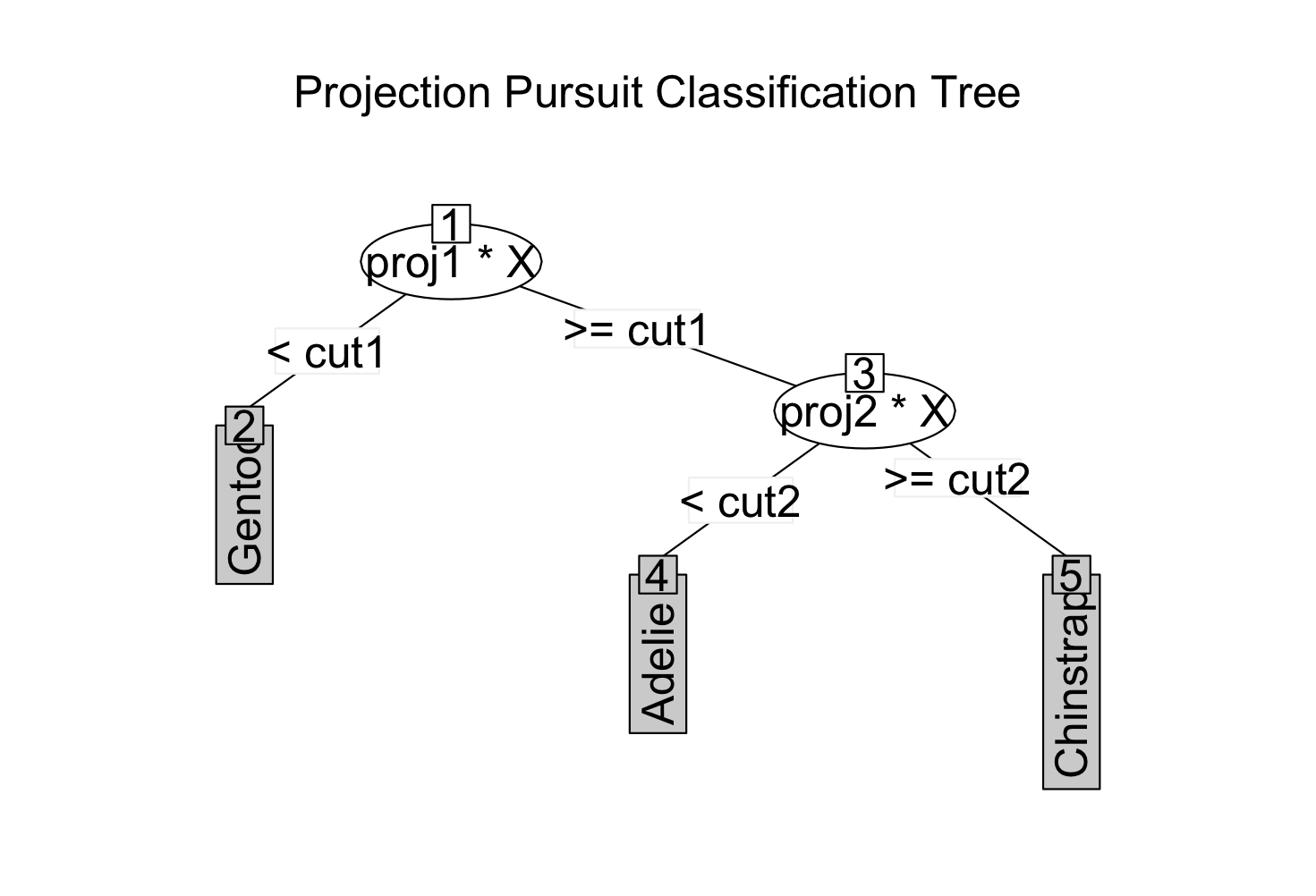
1) root
2)* proj1*X < cut1 -> "Gentoo"
3) proj1*X >= cut1
4)* proj2*X < cut2 -> "Adelie"
5)* proj2*X >= cut2 -> "Chinstrap"
Error rates
-------------------------------------------------------------
[1] 1
pred<- predict(object = penguins_ppt, newdata = penguins_test[,-1], true.class = penguins_test$species)
pred$predict.error
[1] 2
pred$predict.class
[1] "Adelie" "Adelie" "Adelie" "Adelie" "Adelie" "Adelie"
[7] "Adelie" "Adelie" "Adelie" "Adelie" "Adelie" "Adelie"
[13] "Adelie" "Adelie" "Adelie" "Adelie" "Adelie" "Adelie"
[19] "Adelie" "Adelie" "Adelie" "Adelie" "Adelie" "Adelie"
[25] "Adelie" "Adelie" "Adelie" "Adelie" "Adelie" "Adelie"
[31] "Adelie" "Adelie" "Adelie" "Adelie" "Adelie" "Adelie"
[37] "Adelie" "Adelie" "Gentoo" "Gentoo" "Gentoo" "Gentoo"
[43] "Gentoo" "Gentoo" "Gentoo" "Gentoo" "Gentoo" "Gentoo"
[49] "Gentoo" "Gentoo" "Gentoo" "Gentoo" "Gentoo" "Gentoo"
[55] "Gentoo" "Gentoo" "Gentoo" "Gentoo" "Gentoo" "Gentoo"
[61] "Gentoo" "Gentoo" "Gentoo" "Gentoo" "Gentoo" "Gentoo"
[67] "Gentoo" "Gentoo" "Gentoo" "Chinstrap" "Chinstrap" "Chinstrap"
[73] "Chinstrap" "Chinstrap" "Adelie" "Chinstrap" "Chinstrap" "Chinstrap"
[79] "Chinstrap" "Chinstrap" "Chinstrap" "Chinstrap" "Chinstrap" "Adelie"
[85] "Chinstrap" "Chinstrap"plot(penguins_ppt)
These binaries (installable software) and packages are in development.
They may not be fully stable and should be used with caution. We make no claims about them.
Health stats visible at Monitor.